Difference between revisions of "Part:BBa K519010"
| (25 intermediate revisions by 4 users not shown) | |||
| Line 13: | Line 13: | ||
For our iGEM project this year, we have successfully observed that SmtA, expressed under a constitutive promoter (J23109) work very well. We plotted a graph of growth rate by looking at the OD595 every 1 hour, and compared the growth of wild type <i>E. coli</I> and SmtA expressing <I>E. coli</i>. As a result, we found that <i>E. coli</I> expressing SmtA grow better than the WT <i>E. coli</i> at a high Cd(II) concentration. | For our iGEM project this year, we have successfully observed that SmtA, expressed under a constitutive promoter (J23109) work very well. We plotted a graph of growth rate by looking at the OD595 every 1 hour, and compared the growth of wild type <i>E. coli</I> and SmtA expressing <I>E. coli</i>. As a result, we found that <i>E. coli</I> expressing SmtA grow better than the WT <i>E. coli</i> at a high Cd(II) concentration. | ||
| + | |||
| + | |||
| + | ==SCAU-China 2021 iGEM TEAM== | ||
| + | |||
| + | SmtA contains four zinc ions. The protein SmtA contains a cleft lined with Cys-sulfur and His-imidazole ligands that binds four zinc ions in a Zn4Cys9His2 cluster. The four Zn2+ ions and five bridging Cys thiolate sulfurs form two fused six-membered rings with distorted boat conformations [1]. SmtA sequesters and detoxifies four zinc ions per molecule and contains a zinc finger structurally similar to eukaryotic GATA [2]. | ||
| + | |||
| + | Moreover, one of the two Cys coordination zinc ions in SmtA is easy to exchange with the exogenous metal (cadmium)(Fig.1), and the other is inert and hard to replace. | ||
| + | |||
| + | |||
| + | [[File:T--SCAU-China--K4014006.jpg|400px]] | ||
| + | |||
| + | Fig.1. Proton-decoupled one-dimensional 111Cd NMR spectra of SmtA. | ||
| + | |||
| + | |||
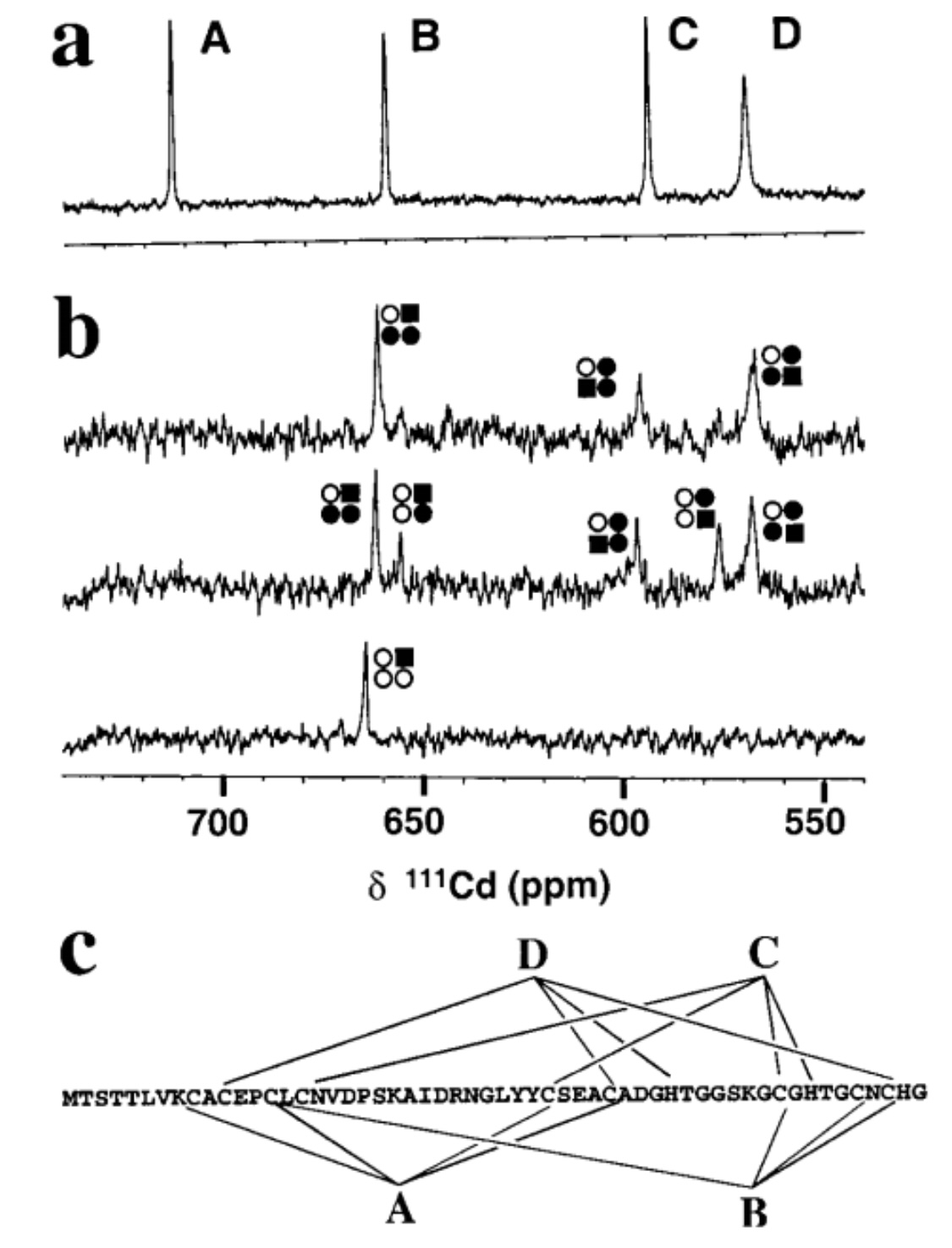
| + | 111Cd4SmtA prepared by reconstitution of apo-SmtA with 111Cd2+ (106.04 MHz,pH 7.0, 318 K, 10% D2O, 50 mM TriszHCl, 50 mM NaCl) showing four peaks with equal integrals. (b) Substitution of Zn2+ by Cd2+. Spectra recorded after mixing Zn4SmtA with 1 (bottom), 4 (middle), and 8 (top) mol equivalents of 111Cd2+ (pH7.0, 308 K, 10% D2O, 50 mM TriszHCl, 50 mM NaCl). Comparison with a shows that the first mol equivalent of 111Cd2+ selectively occupies binding site B, and that binding site A is not occupied at all. The various peaks reflect 111Cd2+ in mixed Cd, Zn clusters. The occupation of the sites is depicted by squares and circles in the order ABDC, clockwise from top left. Filled square, Cd in the site which gives rise to the respective peak; filled circle, Cd in an adjacent site; open circle, Zn; e.g., the peak at 656 ppm corresponds to111Cd2+ in site B, with Zn2+ in sites A and C and 111Cd2+ in site D. (c) | ||
| + | Metal-to-ligand connectivities for SmtA (as deter-mined by 2D heteronuclear NMR experiments) [1]. | ||
| + | |||
| + | In addition, SCAU-China also optimized the codon according to yeast codon bias. The experimental results are shown in BBa_K4014012 | ||
| + | |||
| + | We have improved this part, more details please see | ||
| + | https://parts.igem.org/Part:BBa_K4014012 | ||
| + | |||
| + | Edited by SCAU-China 2021. | ||
| + | |||
| + | ==XJTLU_China 2022 iGEM TEAM== | ||
| + | <html> | ||
| + | Cys in SmtA is the key for the binding of metal ions, such as Cu(II), Cd(II) and Zn(II). Based on the sequence of SmtA, our team mapped out the 3D structure of SmtA and highlighted the position of Cys associated with the metal ion binding sites. | ||
| + | <br> | ||
| + | <br> | ||
| + | <div> | ||
| + | <img src="https://static.igem.wiki/teams/4463/wiki/figures-for-engineering-and-parts/smta-structure.png | ||
| + | " style="width:60%;height=60%; "></img> | ||
| + | </div> | ||
| + | <br> | ||
| + | Figure 1. The predicted structure of SmtA protein and highlighted binding sites. | ||
| + | <br> | ||
| + | <br> | ||
| + | <div> | ||
| + | <img src="https://static.igem.wiki/teams/4463/wiki/figures-for-engineering-and-parts/smta-related.jpeg | ||
| + | " style="width60%;height=60%; "></img> | ||
| + | </div> | ||
| + | <br> | ||
| + | Figure 2. Comparison with non-redundant set of PDB structures. | ||
| + | <br> | ||
| + | For more analysis result, please visit <a href="https://swissmodel.expasy.org/assess/JZc5QC/01">Swiss Model</a>. | ||
| + | <br> | ||
| + | <br> | ||
| + | XJTLU_China also improved the function of SmtA by adding SUMO tag. For more information about the result of improvement, please visit <a href="https://parts.igem.org/Part:BBa_K4463002">BBa_K4463002</a>. | ||
| + | |||
| + | |||
| + | <br> | ||
| + | <br> | ||
| + | </html> | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
===Usage and Biology=== | ===Usage and Biology=== | ||
<P> | <P> | ||
| Line 23: | Line 82: | ||
| − | ===Functional Parameters | + | ===Functional Parameters=== |
| + | <partinfo>BBa_K519010 parameters</partinfo> | ||
| + | ==Functional Parameters: Austin_UTexas== | ||
<html> | <html> | ||
<body> | <body> | ||
| − | + | <h3><center>Burden Imposed by this Part:</center></h3> | |
| − | <h3><center> | + | |
<figure> | <figure> | ||
<div class = "center"> | <div class = "center"> | ||
| Line 36: | Line 96: | ||
</figure> | </figure> | ||
<p> Burden is the percent reduction in the growth rate of <i>E. coli</i> cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the | <p> Burden is the percent reduction in the growth rate of <i>E. coli</i> cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the | ||
| − | <a href="https://parts.igem.org/Part:BBa_K3174002">BBa_K3174002</a> - <a href="https://parts.igem.org/Part:BBa_K3174007">BBa_K3174007</a> pages for more information on the methods and other conclusions from a large-scale measurement project conducted by the <a href=" | + | <a href="https://parts.igem.org/Part:BBa_K3174002">BBa_K3174002</a> - <a href="https://parts.igem.org/Part:BBa_K3174007">BBa_K3174007</a> pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the <a href="http://2019.igem.org/Team:Austin_UTexas">2019 Austin_UTexas team</a>.</p> |
| + | <p>This functional parameter was added by the <a href="https://2020.igem.org/Team:Austin_UTexas/Contribution">2020 Austin_UTexas team.</a></p> | ||
</body> | </body> | ||
</html> | </html> | ||
Latest revision as of 13:03, 11 October 2022
SmtA
SmtA is a metallothionein that can bind to heavy metal ions such as Cd(II). This SmtA is cloned from Synechococcus sp. PCC7942, a cyanobacterial strain. SmtA is a type II metallothionein with a reduced cys content.It was reported that marine cyanobacterium that does not originally harbor a homologous gene, when genetically engineered to express SmtA, could tolerate medium containing Cd(II), Zn(II) and Cu(II) (Sode et al., 1998).

The data above, taken from a paper (Sode et al., 1988), shows that a cyanobacterial strain was able to tolerate medium of high Cd(II) concentration when it harbored the smtA gene.
For our iGEM project this year, we have successfully observed that SmtA, expressed under a constitutive promoter (J23109) work very well. We plotted a graph of growth rate by looking at the OD595 every 1 hour, and compared the growth of wild type E. coli and SmtA expressing E. coli. As a result, we found that E. coli expressing SmtA grow better than the WT E. coli at a high Cd(II) concentration.
SCAU-China 2021 iGEM TEAM
SmtA contains four zinc ions. The protein SmtA contains a cleft lined with Cys-sulfur and His-imidazole ligands that binds four zinc ions in a Zn4Cys9His2 cluster. The four Zn2+ ions and five bridging Cys thiolate sulfurs form two fused six-membered rings with distorted boat conformations [1]. SmtA sequesters and detoxifies four zinc ions per molecule and contains a zinc finger structurally similar to eukaryotic GATA [2].
Moreover, one of the two Cys coordination zinc ions in SmtA is easy to exchange with the exogenous metal (cadmium)(Fig.1), and the other is inert and hard to replace.
Fig.1. Proton-decoupled one-dimensional 111Cd NMR spectra of SmtA.
111Cd4SmtA prepared by reconstitution of apo-SmtA with 111Cd2+ (106.04 MHz,pH 7.0, 318 K, 10% D2O, 50 mM TriszHCl, 50 mM NaCl) showing four peaks with equal integrals. (b) Substitution of Zn2+ by Cd2+. Spectra recorded after mixing Zn4SmtA with 1 (bottom), 4 (middle), and 8 (top) mol equivalents of 111Cd2+ (pH7.0, 308 K, 10% D2O, 50 mM TriszHCl, 50 mM NaCl). Comparison with a shows that the first mol equivalent of 111Cd2+ selectively occupies binding site B, and that binding site A is not occupied at all. The various peaks reflect 111Cd2+ in mixed Cd, Zn clusters. The occupation of the sites is depicted by squares and circles in the order ABDC, clockwise from top left. Filled square, Cd in the site which gives rise to the respective peak; filled circle, Cd in an adjacent site; open circle, Zn; e.g., the peak at 656 ppm corresponds to111Cd2+ in site B, with Zn2+ in sites A and C and 111Cd2+ in site D. (c)
Metal-to-ligand connectivities for SmtA (as deter-mined by 2D heteronuclear NMR experiments) [1].
In addition, SCAU-China also optimized the codon according to yeast codon bias. The experimental results are shown in BBa_K4014012
We have improved this part, more details please see https://parts.igem.org/Part:BBa_K4014012
Edited by SCAU-China 2021.
XJTLU_China 2022 iGEM TEAM
Cys in SmtA is the key for the binding of metal ions, such as Cu(II), Cd(II) and Zn(II). Based on the sequence of SmtA, our team mapped out the 3D structure of SmtA and highlighted the position of Cys associated with the metal ion binding sites.

Figure 1. The predicted structure of SmtA protein and highlighted binding sites.

Figure 2. Comparison with non-redundant set of PDB structures.
For more analysis result, please visit Swiss Model.
XJTLU_China also improved the function of SmtA by adding SUMO tag. For more information about the result of improvement, please visit BBa_K4463002.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal PstI site found at 93
- 12INCOMPATIBLE WITH RFC[12]Illegal PstI site found at 93
- 21COMPATIBLE WITH RFC[21]
- 23INCOMPATIBLE WITH RFC[23]Illegal PstI site found at 93
- 25INCOMPATIBLE WITH RFC[25]Illegal PstI site found at 93
Illegal AgeI site found at 121 - 1000COMPATIBLE WITH RFC[1000]
Functional Parameters
| function | -NA- |
| n/a | SmtA |
Functional Parameters: Austin_UTexas
Burden Imposed by this Part:

Burden is the percent reduction in the growth rate of E. coli cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the BBa_K3174002 - BBa_K3174007 pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the 2019 Austin_UTexas team.
This functional parameter was added by the 2020 Austin_UTexas team.