Difference between revisions of "Part:BBa K2916073"
Konstantinos (Talk | contribs) |
Konstantinos (Talk | contribs) |
||
| (4 intermediate revisions by the same user not shown) | |||
| Line 20: | Line 20: | ||
We tested the functionality of the new part by expressing it in OnePot PURE cell-free system using the <html><a style="padding: 0px; margin: 0px;" href="https://www.protocols.io/view/protein-expression-in-onepot-pure-cell-free-system-8avhse6"> protocol</a></html> in 5μl reactions (with and without triggering DNA) and measuring the excitation at wavelength of 535nm in a platereader at 37°C. We expected the sample without the triggering DNA to produce minimal amount of fluorescence due to unavoidable leakage, while the sample where the triggering DNA is included to be expressed normally. | We tested the functionality of the new part by expressing it in OnePot PURE cell-free system using the <html><a style="padding: 0px; margin: 0px;" href="https://www.protocols.io/view/protein-expression-in-onepot-pure-cell-free-system-8avhse6"> protocol</a></html> in 5μl reactions (with and without triggering DNA) and measuring the excitation at wavelength of 535nm in a platereader at 37°C. We expected the sample without the triggering DNA to produce minimal amount of fluorescence due to unavoidable leakage, while the sample where the triggering DNA is included to be expressed normally. | ||
| + | |||
| + | In all the experiments we performed for our project we used the following sequences, which include a T7 promoter and a disease or grapevine specific sequence, as the triggering DNA: | ||
| + | |||
| + | '''T7_BN_Trigger''': | ||
| + | <br/> | ||
| + | taatacgactcactataggggcaattccaaaagtaaaagcaggtttagcgatggttgtttttcctgaaggtggtattaaagatcgaaatgatgaagcaacggtaccacttttagaggggtctttt | ||
| + | |||
| + | '''T7_EC_Trigger''': | ||
| + | <br/> | ||
| + | taatacgactcactataggaatcctgttttccgaaaacaaccaagggttcagaaaacgataataaaaaaaggataggtgcagagactcaatggaagctgttctaacaaaaacaaatggagttgact | ||
| + | |||
| + | '''T7_FD_Trigger''': | ||
| + | <br/> | ||
| + | taatacgactcactataggttggaggaaggtggggataacgtcaaatcatcatgccccttatgatctgggctacaaacgtgatacaatggctattacaaagagtagctgaaacgcgagtttttag | ||
| + | |||
| + | We used the same triggers for all toeholds in the same family (exp. Trigger BN for all BN toeholds). However for better expression, one should restrict the trigger to the exact complementary sequence to the toehold part of the switch. For each specific toehold switch, the trigger should include a T7 promoter and the complementary sequence of the first 36 nucleotides of the toehold and hairpin region. | ||
| + | |||
| + | |||
<br/> | <br/> | ||
| Line 41: | Line 59: | ||
| − | For more information regarding the design of the toehold please refer to the basic part: <html><a style="padding: 0px; margin: 0px;" href="https://parts.igem.org/Part:BBa_K2916059"> "Toehold for Bois Noir 2.1"</a></html> | + | '''For more information regarding the design of the toehold please refer to the basic part: <html><a style="padding: 0px; margin: 0px;" href="https://parts.igem.org/Part:BBa_K2916059"> "Toehold for Bois Noir 2.1"</a></html> ''' |
Latest revision as of 15:13, 11 October 2020
Toehold switch for Bois Noir 2.1 with superfolder GFP
Summary:
During the development of our project, we decided to improve the "superfolder GFP driven by T7 promoter"
As we wanted to produce a signal only in case of the presence of the desired sequence, we needed to find a way to regulate the protein expression in our system. We achieved that by adding a Toehold, that has an integrated T7 promoter and a Ribosome Binding Site, before the encoded gene sequence. This procedure blocks the expression of sf GFP in absence of trigger DNA and and allows its expression only when the specific sequence is present in our sample.
With our improvement, sf GFP can now be used as a diagnostics tool for detection of Bois Noir disease in the grapevine.
The Toehold switch for Bois Noir 2.1 with superfolder GFP composite part was created with PCR. Specifically, we acquired the existing part in the distribution kit, and removed the promoter and RBS sequence. Then we run PCR with primers that had the sequence of the toehold already build-in.
Hypothesis and method:
We tested the functionality of the new part by expressing it in OnePot PURE cell-free system using the protocol in 5μl reactions (with and without triggering DNA) and measuring the excitation at wavelength of 535nm in a platereader at 37°C. We expected the sample without the triggering DNA to produce minimal amount of fluorescence due to unavoidable leakage, while the sample where the triggering DNA is included to be expressed normally.
In all the experiments we performed for our project we used the following sequences, which include a T7 promoter and a disease or grapevine specific sequence, as the triggering DNA:
T7_BN_Trigger:
taatacgactcactataggggcaattccaaaagtaaaagcaggtttagcgatggttgtttttcctgaaggtggtattaaagatcgaaatgatgaagcaacggtaccacttttagaggggtctttt
T7_EC_Trigger:
taatacgactcactataggaatcctgttttccgaaaacaaccaagggttcagaaaacgataataaaaaaaggataggtgcagagactcaatggaagctgttctaacaaaaacaaatggagttgact
T7_FD_Trigger:
taatacgactcactataggttggaggaaggtggggataacgtcaaatcatcatgccccttatgatctgggctacaaacgtgatacaatggctattacaaagagtagctgaaacgcgagtttttag
We used the same triggers for all toeholds in the same family (exp. Trigger BN for all BN toeholds). However for better expression, one should restrict the trigger to the exact complementary sequence to the toehold part of the switch. For each specific toehold switch, the trigger should include a T7 promoter and the complementary sequence of the first 36 nucleotides of the toehold and hairpin region.
Results:
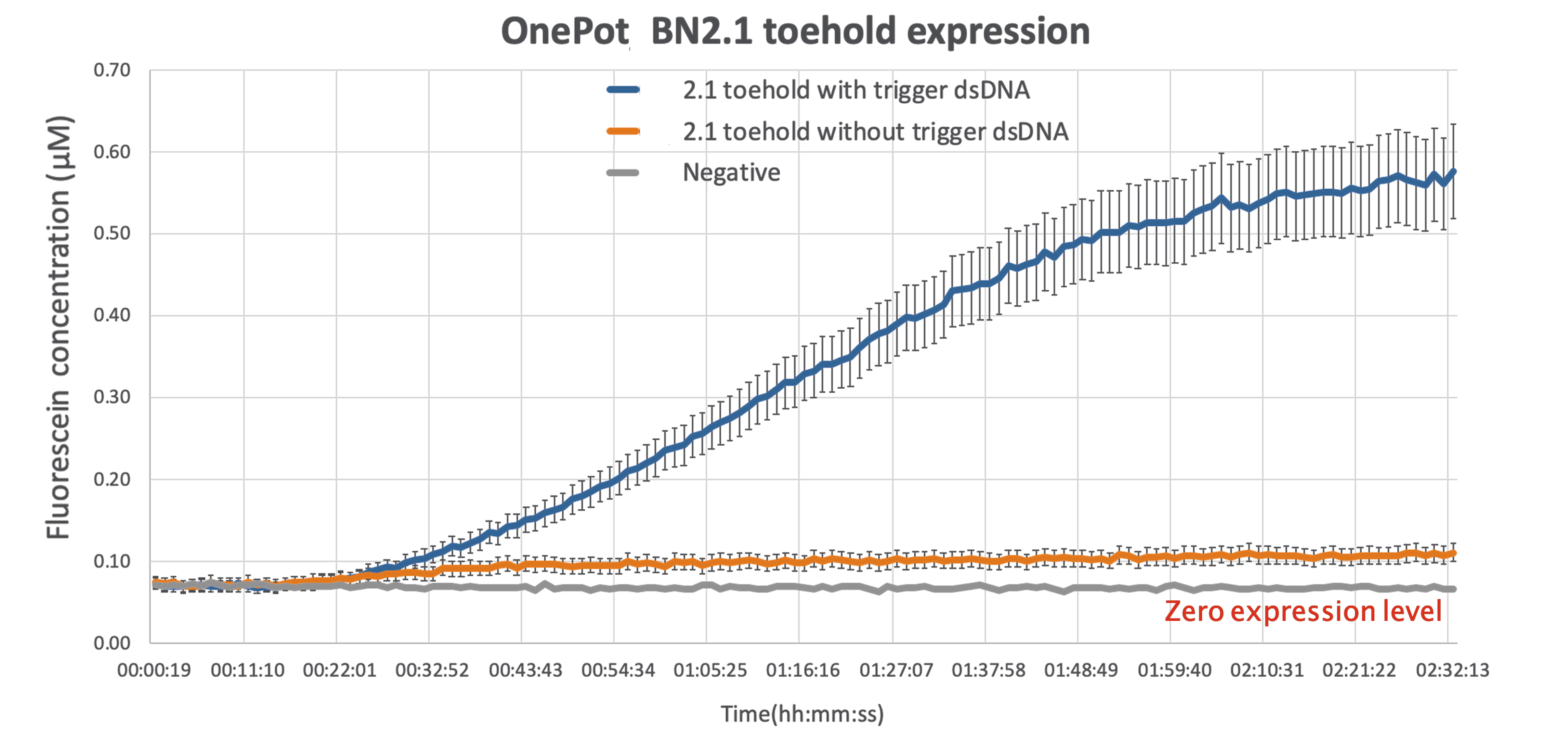
As expected there is some leakage present in the final results,but given that the output of the reading while the wanted sequence is present is more than 5-fold larger, there can be a clear distinction between the ON and OFF phase of the toehold.
This improvement can be act as a proof of concept, demonstrating the ability of sf GFP to be used as a diagnostics tool, simple by modifying the sequence toehold we are using to bind to any desired sequence.
For more information regarding the design of the toehold please refer to the basic part: "Toehold for Bois Noir 2.1"
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 96
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 141
