Difference between revisions of "Part:BBa K3037003"
(→d) Strep-tag column purification) |
(→d) Strep-tag column purification) |
||
| (7 intermediate revisions by the same user not shown) | |||
| Line 40: | Line 40: | ||
The weight of the fusion protein was calculated in accordance to the number of base pairs. The sequence of our Full Construct is 924 bp/3 = 308 amino acids long, each amino acid weights on average 110 Dalton [1], which results in the final weight of approximately 230 kDa. | The weight of the fusion protein was calculated in accordance to the number of base pairs. The sequence of our Full Construct is 924 bp/3 = 308 amino acids long, each amino acid weights on average 110 Dalton [1], which results in the final weight of approximately 230 kDa. | ||
| − | [[File: | + | [[File:T--TU_Dresden--fancyFC.png|center|400px|thumb|none|Figure 1: Visualization of the full construct with its single parts and their respective functions.]] |
| + | |||
== Biology == | == Biology == | ||
| Line 76: | Line 77: | ||
==== 1) Expression of the Full Construct in pOCC97 [https://parts.igem.org/Part:BBa_K3037000 (BBa_K3037000):] monitoring growth of <i>E.coli</i> ==== | ==== 1) Expression of the Full Construct in pOCC97 [https://parts.igem.org/Part:BBa_K3037000 (BBa_K3037000):] monitoring growth of <i>E.coli</i> ==== | ||
| − | Once all the single parts were fused together, the Full Construct was cloned into our expression plasmid pOCC97 [https://parts.igem.org/Part:BBa_K3037000 (BBa_K3037000)]. The correct insertion of our Full Construct into the plasmid was proven via restriction digest followed by agarose gel electrophoresis. For that, we performed a triple digest with PmlI, XbaI and PstI and got several positive clones. This simulation of the digest in SnapGene is depicted in Figure | + | Once all the single parts were fused together, the Full Construct was cloned into our expression plasmid pOCC97 [https://parts.igem.org/Part:BBa_K3037000 (BBa_K3037000)]. The correct insertion of our Full Construct into the plasmid was proven via restriction digest followed by agarose gel electrophoresis. For that, we performed a triple digest with PmlI, XbaI and PstI and got several positive clones. This simulation of the digest in SnapGene is depicted in Figure 2. |
| − | [[File:FC_digestion_gel_final.png|center|400px|thumb|none|Figure | + | [[File:FC_digestion_gel_final.png|center|400px|thumb|none|Figure 2: Multiple Full Construct clones digested with PmlI, XbaI and Pst-1. On the right, the simulation in SnapGene is shown. The positive clones are marked with red crosses.]] |
Furthermore, it was proven that the <span style="font-style: italic;">E. coli</span> could grow normally after the induction of the fusion protein. For this matter, the development of the bacteria cultures was monitored by measuring the OD at 600 nm during different time points before and after induction with 1 mM IPTG. | Furthermore, it was proven that the <span style="font-style: italic;">E. coli</span> could grow normally after the induction of the fusion protein. For this matter, the development of the bacteria cultures was monitored by measuring the OD at 600 nm during different time points before and after induction with 1 mM IPTG. | ||
| − | As shown in Figure | + | As shown in Figure 3, the growth of the bacteria is not affected by the expression of the protein. Important to note, the expression of the Full Construct was performed in two slightly different pOCC97 plasmids, that differ in their Ribosome Binding Site (RBS) and compatibility to the RCF10 BioBrick standard. Hereinafter, they are going to be referred to as optimized and not optimized (read the registry page [https://parts.igem.org/Part:BBa_K3037000 BBa_K3037000] for more details regarding the difference between these two plasmids). |
| − | [[File:T--TU_Dresden--Comparison_optimization_growing_curve_BBa_K3037000.png|center|400px|thumb|none|Figure | + | [[File:T--TU_Dresden--Comparison_optimization_growing_curve_BBa_K3037000.png|center|400px|thumb|none|Figure 3: Comparison of the growth curve compared before and after optimization]] |
| − | To go further, the expression of the Full Construct in pOCC97 at different temperatures was studied. Also, the optimized and not optimized pOOC97 were compared. Both is visualized in Figure | + | To go further, the expression of the Full Construct in pOCC97 at different temperatures was studied. Also, the optimized and not optimized pOOC97 were compared. Both is visualized in Figure 4. |
| − | [[File:Growingcurves.png|center|700px|thumb|none|Figure | + | [[File:Growingcurves.png|center|700px|thumb|none|Figure 4: Comparison of the growth curves of optimized and not optimized pOCC97.]] |
==== 2) SDS-PAGEs for the expression assay over the time of Full Construct [https://parts.igem.org/Part:BBa_K3037003 (BBa_K3037003)] ==== | ==== 2) SDS-PAGEs for the expression assay over the time of Full Construct [https://parts.igem.org/Part:BBa_K3037003 (BBa_K3037003)] ==== | ||
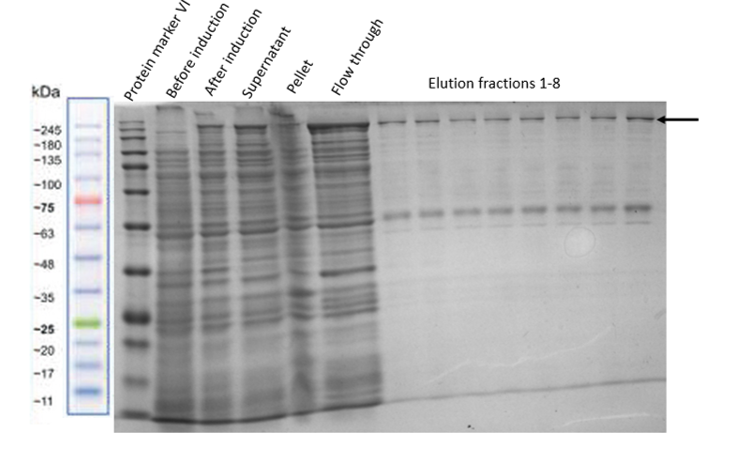
| − | After proving that the final construct was well inserted in our plasmid, it was expressed overnight. The first expression, performed at 37°C for seven hours was induced with 1 mM IPTG. The result is shown in Figure | + | After proving that the final construct was well inserted in our plasmid, it was expressed overnight. The first expression, performed at 37°C for seven hours was induced with 1 mM IPTG. The result is shown in Figure 5: |
| − | [[File:FC_expression.png|center|400px|thumb|none|Figure | + | [[File:FC_expression.png|center|400px|thumb|none|Figure 5: SDS-page showing the expression of the Full Construct in pOCC97 after induction with 1 mM IPTG. Different type points show the increased expression of the Full Construct (marked with black arrow).]] |
| − | To compare the best expression conditions, the same experiment was repeated several times at different temperatures and IPTG concentrations in both plasmids. The results are shown in Figures | + | To compare the best expression conditions, the same experiment was repeated several times at different temperatures and IPTG concentrations in both plasmids. The results are shown in Figures 6 to 8. |
<b>Expression of Full Construct in pOCC97 not optimized at 18ºC and different IPTG concentrations</b> | <b>Expression of Full Construct in pOCC97 not optimized at 18ºC and different IPTG concentrations</b> | ||
| − | [[File:T--TU_Dresden--Expression_FullConstruct_notoptimized_18_BBa_K3037000.png|center|400px|thumb|none|Figure | + | [[File:T--TU_Dresden--Expression_FullConstruct_notoptimized_18_BBa_K3037000.png|center|400px|thumb|none|Figure 6: Expression of the Full Construct in not optimized pOCC97 at 18ºC. The timepoints 0 to 3 relate to 1,2,3 and 10 h post induction respectively. IPTG was added in concentrations of 0.2 nmM, 0.5 mM and 1 mM. The black arrow is corresponding to the band at the expected protein size of 230 kDa.]] |
<b>Expression of Full Construct in pOCC97 not optimized at 37ºC and different IPTG concentrations</b> | <b>Expression of Full Construct in pOCC97 not optimized at 37ºC and different IPTG concentrations</b> | ||
| − | [[File:T--TU_Dresden--Expression_FullConstruct_notoptimized_37.2_BBa_K3037000.png|center|400px|thumb|none|Figure | + | [[File:T--TU_Dresden--Expression_FullConstruct_notoptimized_37.2_BBa_K3037000.png|center|400px|thumb|none|Figure 7: Expression of the Full Construct in not optimized pOCC97 at 37ºC. The timepoints 0 to 3 relate to 1,2,3 and 10 h post induction respectively. IPTG was added in concentrations of 0.2 nmM, 0.5 mM and 1 mM. The black arrow is corresponding to the band at the expected protein size of 230 kDa.]] |
<b>Expression of the Full Construct in pOCC97 optimized at different temperatures and IPTG concentrations</b> | <b>Expression of the Full Construct in pOCC97 optimized at different temperatures and IPTG concentrations</b> | ||
| − | [[File:T--TU_Dresden--Expression_FullConstruct_optimized_BBa_K3037000.png|center|400px|thumb|none|Figure | + | [[File:T--TU_Dresden--Expression_FullConstruct_optimized_BBa_K3037000.png|center|400px|thumb|none|Figure 8: Expression of the Full Construct in optimized pOCC97 at different temperatures and IPTG concentrations. The timepoints 0 to 3 relate to 1,2,3 and 10 h post induction respectively. IPTG was added in concentrations of either 0.5 mM or 1 mM. The cutures were grown at 37°C or 18°C as indicated in the figure. The black arrow is corresponding to the band at the expected protein size of 230 kDa.]] |
==== 3) Image analysis of the expression in the SDS-PAGEs with ImageJ ==== | ==== 3) Image analysis of the expression in the SDS-PAGEs with ImageJ ==== | ||
| Line 115: | Line 116: | ||
<b>Temperature and IPTG induction dependence of the optimized pOCC97</b> | <b>Temperature and IPTG induction dependence of the optimized pOCC97</b> | ||
| − | [[File:Clone7_dependences.png|center|600px|thumb|none|Figure | + | [[File:Clone7_dependences.png|center|600px|thumb|none|Figure 9: Expression of the Full Construct in optimized pOCC97 under different conditions.]] |
| − | Figure | + | Figure 9 indicates that at 18°C an induction with 0.5 mM IPTG results in higher yields in the optimized pOCC97 compared to an induction with 1 mM. Even if at 37°C the amount of protein expression rises faster, after overnight incubation more protein is produced in the 18°C culture. |
<b>Temperature and IPTG induction dependence of the not optimized pOCC97</b> | <b>Temperature and IPTG induction dependence of the not optimized pOCC97</b> | ||
| − | [[File:Clone11_dependences.png|center|600px|thumb|none|Figure | + | [[File:Clone11_dependences.png|center|600px|thumb|none|Figure 10: Expression of the Full Construct in not optimized pOCC97 under different conditions.]] |
The experiments for the not optimized pOCC97 showed a different preference, here the yields are if grown at 37°C unless induction is done with 1 mM IPTG. | The experiments for the not optimized pOCC97 showed a different preference, here the yields are if grown at 37°C unless induction is done with 1 mM IPTG. | ||
<b>Comparison between optimized and not optimized pOCC97</b> | <b>Comparison between optimized and not optimized pOCC97</b> | ||
| − | [[File:Comparison_both.png|center|600px|thumb|none|Figure | + | [[File:Comparison_both.png|center|600px|thumb|none|Figure 11: Comparison between the expression of optimized and not optimized pOCC97.]] |
| − | Based on the analysis of Figures | + | Based on the analysis of Figures 9 to 11 it can be concluded that the optimal conditions for the expression of [https://parts.igem.org/Part:BBa_K3037003 BBa_3037003] in pOCC97 are 18ºC and 0.5 mM IPTG. The expression seems to be more stable over time for the optimized plasmid than for the non optimized. |
==== 4) Characterization of the single parts of the Full Construct ==== | ==== 4) Characterization of the single parts of the Full Construct ==== | ||
| Line 132: | Line 133: | ||
===== a) Purification via MBP ===== | ===== a) Purification via MBP ===== | ||
| − | After ensuring that the Full Construct is expressed properly in our plasmid by improving its expression conditions, it was purified by using amylose resin to bind its MBP site. To test for the correct functioning of the MBP-tag of the fusion protein we performed different experiments. For that, two different protocols were used. On the one hand, an amylose resin column was used, and on the other hand, a batch binding solution was prepared. Better results were obtained with the latter one. For the batch binding, the resin was pipetted into a falcon and incubated with the cell lysate for 1.5 hours on a rotator at 4°C. The SDS-PAGE of the purification steps is shown in Figure | + | After ensuring that the Full Construct is expressed properly in our plasmid by improving its expression conditions, it was purified by using amylose resin to bind its MBP site. To test for the correct functioning of the MBP-tag of the fusion protein we performed different experiments. For that, two different protocols were used. On the one hand, an amylose resin column was used, and on the other hand, a batch binding solution was prepared. Better results were obtained with the latter one. For the batch binding, the resin was pipetted into a falcon and incubated with the cell lysate for 1.5 hours on a rotator at 4°C. The SDS-PAGE of the purification steps is shown in Figure 12. |
| − | [[File:MBP_batchbinding.png|center|400px|thumb|none|Figure | + | [[File:MBP_batchbinding.png|center|400px|thumb|none|Figure 12: SDS-PAGE of purification of Full Construct (indicated by the black arrow) with MBP-tag and an amylose resin batch binding step.]] |
===== b) Activity assay of HRP ===== | ===== b) Activity assay of HRP ===== | ||
| Line 144: | Line 145: | ||
[[File:Cuvetten_mod.mp4]] | [[File:Cuvetten_mod.mp4]] | ||
| − | To further verify the correct activity of the HRP, the absorbance at 650 nm of both cell lysates was measured. As it can be seen in Figure | + | To further verify the correct activity of the HRP, the absorbance at 650 nm of both cell lysates was measured. As it can be seen in Figure 13, the cell lysate that is expressing MBP showed a much lower absorbance at 650 nm, than the lysate containing the Full Construct. This means, that the HRP contained in Full Construct is working properly. |
| − | [[File:Cell_lysate_Full_Construct.png|center|300px|thumb|none|Figure | + | [[File:Cell_lysate_Full_Construct.png|center|300px|thumb|none|Figure 13: Absorbance measured at 650 nm of cell lysates with and without Full Construct after TMB addition.]] |
===== c) dCas9 activity===== | ===== c) dCas9 activity===== | ||
| + | The DNA binding activity of the dCas9 contained in our Full Construct was proven via EMSA. Our dCas9 is expected to bind to the <i>sry</i> gene, since the guide RNA was specifically designed to target this gene. In Figure 14, it can be clearly seen how at very high concentrations of expressed Full Construct, our dCas9 is able to completely bind to the <i>sry</i> gene, fully hindering its mobility through the gel (red box). | ||
| + | |||
| + | [[File:T--TU_Dresden--emsa1.png|center|400px|thumb|none|Figure 14: Proof of the binding of our Full Construct (with its dCas9) to the DNA sequence of interest (SRY gene in this experiment).]] | ||
| + | |||
| + | |||
| + | In order to determine the lowest concentration at which our expressed Full Construct causes the pull of the gene, different concentrations were loaded on the gradient TBE acrylamide gel. We found that at approximately 1.28 ug the dCas9 is able to bind and therefore, pull up the DNA. Additionally, at 8.56 ug we can see a very clear shift of the DNA, since it can be seen in the region marked with a red box in Figure 15. | ||
| + | |||
| + | [[File:T--TU_Dresden--emsa2.png|center|600px|thumb|none|Figure 15: Determination of the lowest concentration of the Full Construct (with dCas9) needed to bind and pull DNA.]] | ||
===== d) Strep-tag column purification ===== | ===== d) Strep-tag column purification ===== | ||
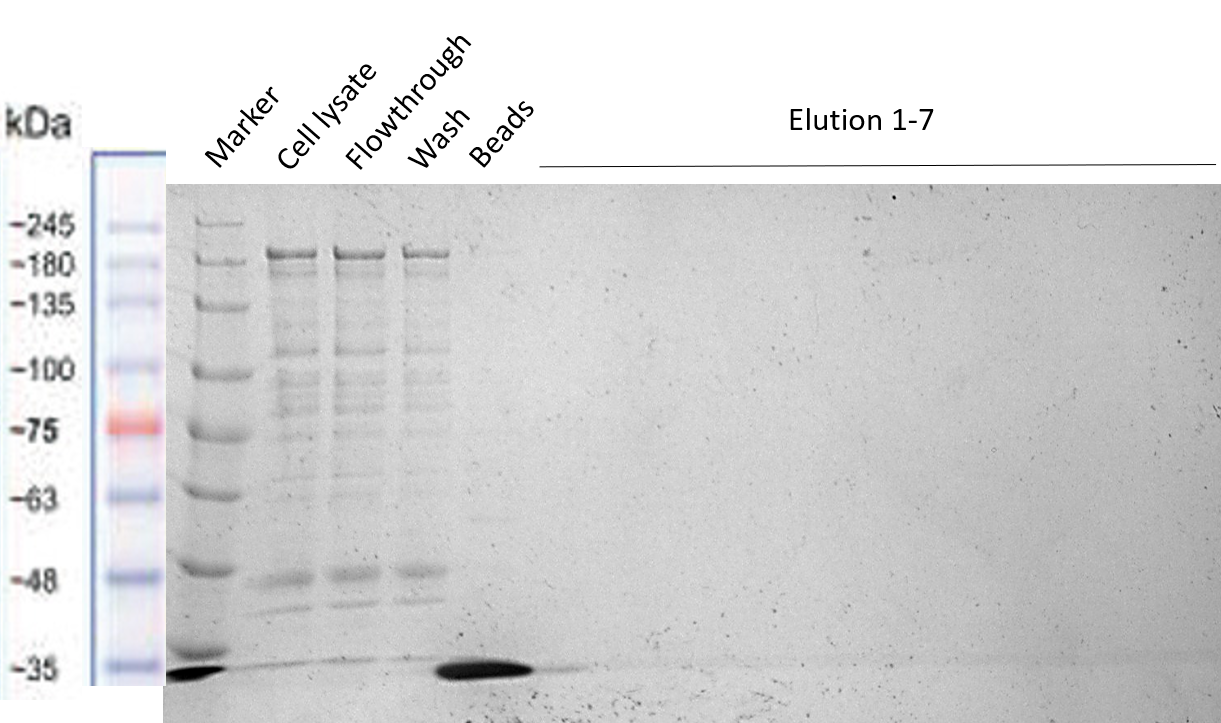
| − | The reason to include a Strep-tag at the end of our Full Construct was to facilitate its purification. However, as already explained in the Registry page of the Strep-tag itself [https://parts.igem.org/Part:BBa_K823038 BBa_K823038,] this BioBrick seems to not be working properly for column purification. The Strep-tag was developed to be used for Western Blots and not for column purification. That is why the purification via Strep-tag did not work (see Figure | + | The reason to include a Strep-tag at the end of our Full Construct was to facilitate its purification. However, as already explained in the Registry page of the Strep-tag itself [https://parts.igem.org/Part:BBa_K823038 BBa_K823038,] this BioBrick seems to not be working properly for column purification. The Strep-tag was developed to be used for Western Blots and not for column purification. That is why the purification via Strep-tag did not work (see Figure 16). However, it was shown in the section before, that we were able to successfully purify our Full Construct via the MBP. |
| − | [[File:Strep_purification_2.png|center|400px|thumb|none|Figure | + | [[File:Strep_purification_2.png|center|400px|thumb|none|Figure 16: Different Full Construct samples before, while and after Strep purification]] |
== Sequence == | == Sequence == | ||
Latest revision as of 02:50, 22 October 2019
Fusion protein dCas9 + HRP (MBP/dCas9/linker/HRP/Strep-tag)
| Fusion protein | |
|---|---|
| Function | Colour detection of specific DNA sequences |
| Use in | Escherichia coli |
| RFC standard | Freiburg RFC25 standard |
| Backbone | pSB1C3 |
| Experimental Backbone | pOCC97 |
| Submitted by | Team: TU_Dresden 2019 |
Contents
- 1 Overview
- 2 Biology
- 3 Characterization
- 3.1 Outline
- 3.2 Experiments in Detail
- 3.2.1 1) Expression of the Full Construct in pOCC97 (BBa_K3037000): monitoring growth of E.coli
- 3.2.2 2) SDS-PAGEs for the expression assay over the time of Full Construct (BBa_K3037003)
- 3.2.3 3) Image analysis of the expression in the SDS-PAGEs with ImageJ
- 3.2.4 4) Characterization of the single parts of the Full Construct
- 4 Sequence
- 5 References
Overview
The TU Dresden 2019 team has designed this BioBrick in order to allow for the quick detection of specific DNA sequences of interest (more information).
The Full Construct as it is shown in Figure 1 below, contains five individual parts each of which have been optimized for it. The MBP and Strep-tag allow purification via amylose resin and Strep-columns, respectively. Additionally, the MBP enhances the expression of dCas9-fusion proteins, and the linker aids for the folding process. The dCas9 identifies the sequence of interest (with the help of an added guideRNA), and the HRP provides with an easily detectable amplified color readout. The single constructs were cloned into pSB1C3 and pSB1A3, and finally, the full construct was inserted into the BBa_K3037000 vector for expression and characterization in Escherichia coli (E. coli).
The weight of the fusion protein was calculated in accordance to the number of base pairs. The sequence of our Full Construct is 924 bp/3 = 308 amino acids long, each amino acid weights on average 110 Dalton [1], which results in the final weight of approximately 230 kDa.
Biology
For more information regarding the biology, design and function of each of the basic parts, please check the individual registry pages:
- MBP: BBa_K3037001
- dCas9: BBa_K3037002
- Linker: BBa_K3037004
- HRP: BBa_K3037007
- Strep-tag: BBa_K823038
The expected parameters of the new protein (determined with ExPASy ProtParam tool) were determined to be:
- Extinction coefficient: 205750 L/(mol*cm)
- Estimated half-life > 30 hours in mammalian cells, >20 hours in yeast and >10 hours in E. coli
Characterization
Outline
We performed the following characterization experiments:
1) Expression of the Full Construct in pOCC97 (BBa_K3037000): monitoring growth of E.coli
2) SDS-PAGEs for the expression assay over the time of Full Construct (BBa_K3037003)
3) Image analysis of the expression in the SDS-PAGEs with ImageJ
4) Characterization of the single parts of the Full Construct
Experiments in Detail
1) Expression of the Full Construct in pOCC97 (BBa_K3037000): monitoring growth of E.coli
Once all the single parts were fused together, the Full Construct was cloned into our expression plasmid pOCC97 (BBa_K3037000). The correct insertion of our Full Construct into the plasmid was proven via restriction digest followed by agarose gel electrophoresis. For that, we performed a triple digest with PmlI, XbaI and PstI and got several positive clones. This simulation of the digest in SnapGene is depicted in Figure 2.
Furthermore, it was proven that the E. coli could grow normally after the induction of the fusion protein. For this matter, the development of the bacteria cultures was monitored by measuring the OD at 600 nm during different time points before and after induction with 1 mM IPTG.
As shown in Figure 3, the growth of the bacteria is not affected by the expression of the protein. Important to note, the expression of the Full Construct was performed in two slightly different pOCC97 plasmids, that differ in their Ribosome Binding Site (RBS) and compatibility to the RCF10 BioBrick standard. Hereinafter, they are going to be referred to as optimized and not optimized (read the registry page BBa_K3037000 for more details regarding the difference between these two plasmids).
To go further, the expression of the Full Construct in pOCC97 at different temperatures was studied. Also, the optimized and not optimized pOOC97 were compared. Both is visualized in Figure 4.
2) SDS-PAGEs for the expression assay over the time of Full Construct (BBa_K3037003)
After proving that the final construct was well inserted in our plasmid, it was expressed overnight. The first expression, performed at 37°C for seven hours was induced with 1 mM IPTG. The result is shown in Figure 5:
To compare the best expression conditions, the same experiment was repeated several times at different temperatures and IPTG concentrations in both plasmids. The results are shown in Figures 6 to 8.
Expression of Full Construct in pOCC97 not optimized at 18ºC and different IPTG concentrations
Expression of Full Construct in pOCC97 not optimized at 37ºC and different IPTG concentrations
Expression of the Full Construct in pOCC97 optimized at different temperatures and IPTG concentrations

3) Image analysis of the expression in the SDS-PAGEs with ImageJ
The previously shown SDS-PAGEs were further analysed by using the software ImageJ to correct for loading differences and to be able to draw conclusions about the best conditions to express the Full Construct in pOCC97.
Temperature and IPTG induction dependence of the optimized pOCC97
Figure 9 indicates that at 18°C an induction with 0.5 mM IPTG results in higher yields in the optimized pOCC97 compared to an induction with 1 mM. Even if at 37°C the amount of protein expression rises faster, after overnight incubation more protein is produced in the 18°C culture.
Temperature and IPTG induction dependence of the not optimized pOCC97
The experiments for the not optimized pOCC97 showed a different preference, here the yields are if grown at 37°C unless induction is done with 1 mM IPTG.
Comparison between optimized and not optimized pOCC97
Based on the analysis of Figures 9 to 11 it can be concluded that the optimal conditions for the expression of BBa_3037003 in pOCC97 are 18ºC and 0.5 mM IPTG. The expression seems to be more stable over time for the optimized plasmid than for the non optimized.
4) Characterization of the single parts of the Full Construct
a) Purification via MBP
After ensuring that the Full Construct is expressed properly in our plasmid by improving its expression conditions, it was purified by using amylose resin to bind its MBP site. To test for the correct functioning of the MBP-tag of the fusion protein we performed different experiments. For that, two different protocols were used. On the one hand, an amylose resin column was used, and on the other hand, a batch binding solution was prepared. Better results were obtained with the latter one. For the batch binding, the resin was pipetted into a falcon and incubated with the cell lysate for 1.5 hours on a rotator at 4°C. The SDS-PAGE of the purification steps is shown in Figure 12.
b) Activity assay of HRP
The inverstigation of the activity of HRP in the Full Construct was done in a dynamic assay. The absorbance at 650 nm was measured over time with the substrate TMB. TMB is a colorless solution but is converted into a blue product by oxidation through HRP. Furthermore, the addition of hydrogen peroxide catalyses the oxidation of the TMB. It acts as a electron donor, enhancing the formation of the blue product. The reaction can be stopped by adding an acidic solution (for example: HCl), resulting in a yellow coloured-readout. (See more information about the HRP activity in BBa_K1800002) To prove the correct activity of the HRP in our Full Construct we performed mechanical lysis on our E.coli cells that were expressing our fusion protein, added the HRP substrate TMB followed by the addition of hydrogen peroxide and stopped the reaction by adding HCl. E.coli expressing our MBP BioBrick were used as a negative control. The result can be seen very nicely in the following video:
To further verify the correct activity of the HRP, the absorbance at 650 nm of both cell lysates was measured. As it can be seen in Figure 13, the cell lysate that is expressing MBP showed a much lower absorbance at 650 nm, than the lysate containing the Full Construct. This means, that the HRP contained in Full Construct is working properly.
c) dCas9 activity
The DNA binding activity of the dCas9 contained in our Full Construct was proven via EMSA. Our dCas9 is expected to bind to the sry gene, since the guide RNA was specifically designed to target this gene. In Figure 14, it can be clearly seen how at very high concentrations of expressed Full Construct, our dCas9 is able to completely bind to the sry gene, fully hindering its mobility through the gel (red box).
In order to determine the lowest concentration at which our expressed Full Construct causes the pull of the gene, different concentrations were loaded on the gradient TBE acrylamide gel. We found that at approximately 1.28 ug the dCas9 is able to bind and therefore, pull up the DNA. Additionally, at 8.56 ug we can see a very clear shift of the DNA, since it can be seen in the region marked with a red box in Figure 15.
d) Strep-tag column purification
The reason to include a Strep-tag at the end of our Full Construct was to facilitate its purification. However, as already explained in the Registry page of the Strep-tag itself BBa_K823038, this BioBrick seems to not be working properly for column purification. The Strep-tag was developed to be used for Western Blots and not for column purification. That is why the purification via Strep-tag did not work (see Figure 16). However, it was shown in the section before, that we were able to successfully purify our Full Construct via the MBP.
Sequence
NOTE: Please be aware, that by combining all the basic parts used for this composite part, the registry automatically inserted a RFC23 scar. However, the design and our cloning strategy is based on RFC25.
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 2302
Illegal NheI site found at 5500 - 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 381
Illegal BglII site found at 5742
Illegal BamHI site found at 4581
Illegal BamHI site found at 5314
Illegal XhoI site found at 5826 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 79