Difference between revisions of "Part:BBa K2927005"
WeiChengWang (Talk | contribs) |
(→New Hair-pin reporter with enhanced trans-cleavage activity of Cas12a.) |
||
| (21 intermediate revisions by 7 users not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K2927005 short</partinfo> | <partinfo>BBa_K2927005 short</partinfo> | ||
| − | CRISPR Cas12a system is one of the bacterial adaptive immune systems. Cas12a protein is the RNA-guided enzyme that binds and cut DNA. When Cas12a protein bind with the specific crRNA, it will be activated. After Cas12a protein is activated, it will cut the target DNA as well as non-specific single-strand DNA (ssDNA), this certain function is used in our project. We designed six crRNA which is derived from the African swine fever virus (ASFV) to detect the DNA of the virus. This sequence is just a part of DNA sequence of the vp72 membrane protein of ASFV. | + | CRISPR Cas12a system is one of the bacterial adaptive immune systems. Cas12a protein is the RNA-guided enzyme that binds and cut DNA. When Cas12a protein bind with the specific crRNA, it will be activated. After Cas12a protein is activated, it will cut the target DNA as well as non-specific single-strand DNA (ssDNA), this certain function is used in our project. We designed six crRNA which is derived from the African swine fever virus (ASFV) to detect the DNA of the virus. This sequence is just a part of DNA sequence of the vp72 membrane protein of ASFV. |
'''Reference'''<br> | '''Reference'''<br> | ||
| − | Janice S. Chen, Enbo Ma, Lucas B. Harrington, | + | Janice S. Chen, Enbo Ma, Lucas B. Harrington, Maria Da Costa, Xinran Tian, Joel M. Palefsky, Jennifer A. Doudna, Chen et al., Science 360, 436–439 (2018) |
| Line 12: | Line 12: | ||
*Introduction | *Introduction | ||
In our project, we combined three parts of biological reactions to detect ASFV specific sequence in samples. The first one is LbCas12a-crRNA system, which can specifically recognize ASFV specific double stranded DNA (dsDNA) sequence on P72 gene. The secondary part is the trans-activation of LbCas12a-crRNA system. When LbCas12a-crRNA system binds to ASFV specific dsDNA sequences, LbCas12a-crRNA system will cleave dsDNA and further degrade non-specific single stranded DNA (ssDNA). To detect the degradation of ssDNA in ASFV-activated LbCas12a-crRNA system, we will use the PicoGreen fluoresce dye to monitor the undegraded ssDNA, which is the third part. To transfer reaction from part I/II/III to detection, we plane to conjugate ssDNA on magnetic beads. The ssDNA conjugated magnetic beads will be easily captured and transfer by electromagnetic force. In the following result section, we will show our progress through experiments that supported our project design. | In our project, we combined three parts of biological reactions to detect ASFV specific sequence in samples. The first one is LbCas12a-crRNA system, which can specifically recognize ASFV specific double stranded DNA (dsDNA) sequence on P72 gene. The secondary part is the trans-activation of LbCas12a-crRNA system. When LbCas12a-crRNA system binds to ASFV specific dsDNA sequences, LbCas12a-crRNA system will cleave dsDNA and further degrade non-specific single stranded DNA (ssDNA). To detect the degradation of ssDNA in ASFV-activated LbCas12a-crRNA system, we will use the PicoGreen fluoresce dye to monitor the undegraded ssDNA, which is the third part. To transfer reaction from part I/II/III to detection, we plane to conjugate ssDNA on magnetic beads. The ssDNA conjugated magnetic beads will be easily captured and transfer by electromagnetic force. In the following result section, we will show our progress through experiments that supported our project design. | ||
| + | |||
*Our targets | *Our targets | ||
Steps to establish CRISPR-LbCas12a system | Steps to establish CRISPR-LbCas12a system | ||
| − | + | '''Expression of LbCas12a protein:''' | |
We transformed pHMT-LbCas12a into E.coli BL21, and then added 0.2 mM IPTG to induce protein expression (see notebook for details). The result showed that the LbCas12a protein expression in soluble fraction was induced by IPTG, and increased as time goes on (Figure 1). The predicted protein size of LbCas12a with MBP and His-tag is about 180 kDa, which is close to the induced protein indicted by red arrow in figure 1. We also examined the insoluble fraction of IPTG induced BL21 by SDS-PAGE, and confirmed that most LbCas12a protein was soluble (Figure 2). | We transformed pHMT-LbCas12a into E.coli BL21, and then added 0.2 mM IPTG to induce protein expression (see notebook for details). The result showed that the LbCas12a protein expression in soluble fraction was induced by IPTG, and increased as time goes on (Figure 1). The predicted protein size of LbCas12a with MBP and His-tag is about 180 kDa, which is close to the induced protein indicted by red arrow in figure 1. We also examined the insoluble fraction of IPTG induced BL21 by SDS-PAGE, and confirmed that most LbCas12a protein was soluble (Figure 2). | ||
| − | [[Image:T--CCU_Taiwan--protein-result-figure1.jpg| | + | [[Image:T--CCU_Taiwan--protein-result-figure1.jpg|500px|thumb|center|'''Fig 1. The Coomassie Blue staining of total protein expression in supernatant of BL21 lysate at different time points.'''Lane 1: Protein Marker (SMOBIO PM2600) ; lane 2: supernatant of BL21 without IPTG induction; lane 3 ~ 10: supernatant of BL21 induced by 0.2 mM IPTG at 16 °C for 16 hr |
| − | + | ||
| − | Lane 1: Protein Marker (SMOBIO PM2600) ; lane 2: supernatant of BL21 without IPTG induction; lane 3 ~ 10: supernatant of BL21 induced by 0.2 mM IPTG at 16 °C for 16 hr | + | |
]] | ]] | ||
| + | [[Image:T--CCU Taiwan--protein-result-figure2.jpg|500px|thumb|center|'''Fig 2. The Coomassie blue staining of total protein expression in pellet of BL21 lysate at different time points.''' | ||
| + | Lane 1: Protein Marker (SMOBIO PM2600); lane 2: pellet of cell lysate without IPTG induction; lane 3 ~ 10: pellet of cell lysate induced by 0.2 mM IPTG at 16 °C for 16 hr.]] | ||
| − | + | ||
| + | '''Pre-test of LbCas12a protein purification:''' | ||
After confirming the induction of LbCas12a protein expression, we purify LbCas12a protein by Ni2+-magnetic beads to pull down the His-tag on LbCas12a protein from soluble fraction. The elution of LbCas12a protein from Ni2+-magnetic beads by excess imidazole or TEV enzyme digestion further confirmed that the Ni2+-magnetic beads purification is clear and easy to reverse (Figure 3). | After confirming the induction of LbCas12a protein expression, we purify LbCas12a protein by Ni2+-magnetic beads to pull down the His-tag on LbCas12a protein from soluble fraction. The elution of LbCas12a protein from Ni2+-magnetic beads by excess imidazole or TEV enzyme digestion further confirmed that the Ni2+-magnetic beads purification is clear and easy to reverse (Figure 3). | ||
| − | '''According to the experiments above, we can express and purified LbCas12a proteins for further application. Therefore we scale up the expression and purification of LbCas12a protein. | + | [[Image:T--CCU Taiwan--protein-result-figure3.jpg |500px|thumb|center|'''Fig 3. The Coomassie blue staining of protein purification by Ni2+-magnet beads.''' |
| + | Lysate: supernatant of cell lysate; flow-through: supernatant before resuspension with Ni2+-magnet beads; TEV treated: treatment of Ni2+-magnet bead-LbCas12a complex by TEV enzyme. | ||
| + | ]] | ||
| + | |||
| + | |||
| + | According to the experiments above, we can express and purified LbCas12a proteins for further application. Therefore we scale up the expression and purification of LbCas12a protein. | ||
| + | |||
| − | + | '''Large scale protein purification:''' | |
We use immobilized metal affinity chromatography (IMAC) to purify LbCas12a protein from soluble fraction of BL21 by Ni2+ chelating sepharose column. We then elute LbCas12a protein from Ni2+ column by imidazole and subjected into FPLC separation. The absorption peak at 27-30 fractions was indicated by red arrow in figure 4. | We use immobilized metal affinity chromatography (IMAC) to purify LbCas12a protein from soluble fraction of BL21 by Ni2+ chelating sepharose column. We then elute LbCas12a protein from Ni2+ column by imidazole and subjected into FPLC separation. The absorption peak at 27-30 fractions was indicated by red arrow in figure 4. | ||
To further confirm that the absorption peak, we performed SDS-PAGE and Coomassie blue staining to fractions 27-30, showing that the absorption peak is indeed LbCas12a protein. | To further confirm that the absorption peak, we performed SDS-PAGE and Coomassie blue staining to fractions 27-30, showing that the absorption peak is indeed LbCas12a protein. | ||
| + | [[Image:T--CCU Taiwan--protein-result-figure4.jpg |500px|thumb|center|'''Fig 4. Chromatogram of LbCas12a purification by Ni2+ chelating sepharose column.''' | ||
| + | The numbers shown in red indicate the fractions. Each fraction collect 5 ml of wash or elute. | ||
| + | ]] | ||
| + | [[Image:T--CCU Taiwan--protein-result-figure5.jpg |500px|thumb|center|'''Fig.5. The Coomassie blue staining of elution analyzed by SDS-PAGE.''' | ||
| + | M: marker; L: lysate; FT: flow-through; number: number of fraction tube. | ||
| + | ]] | ||
| − | <!-- Add more about the biology of this part here | + | <!-- Add more about the biology of this part here--> |
| − | ===Usage | + | |
| + | ===<html><a href="https://2021.igem.org/Team:UPF_Barcelona" target="_blank" onmouseover="this.style.color='#ff724a';" onmouseout="this.style.color='#ff9515';" style="color: #ff9515; text-decoration: none; margin-right:-15px;">UPF Barcelona ARIA Team </a></html>experimental application=== | ||
| + | |||
| + | '''Usage, Biology and Characterization''' | ||
| + | <br> | ||
| + | ARIA’s biosensors are based on two main elements: the endonuclease enzyme LbCas12a from Lachnospiraceae Bacterium, and the crRNAs designed specifically to target antibiotic resistance (AR) sequences. Once the two elements bind and the target is detected, a fluorescent signal is activated due to LbCas12a cis cleavage activity. However, these elements have been implemented on living E.Coli (BL21), and become active after lysing the cells and being put in contact with AR sequences. This has given rise to a new innovative approach: the in vivo – in vitro detection. | ||
| + | |||
| + | To do so, the original plasmid employed for cloning Cas12 was taken from (<html><a href="https://www.addgene.org/113431/" target="_blank" onmouseover="this.style.color='#ff724a';" onmouseout="this.style.color='#ff9515';" style="color: #ff9515; text-decoration: none; margin-right:-15px;">Addgene</a></html>), and it was optimized by deleting purification sites and unuseful sequences codifying for MBP (Maltose Binding Protein). Cas12 is expressed under the T7 promoter so that it is inducible with IPTG, and it has ampicillin resistance to allow for its selection. | ||
| + | |||
| + | 5 gRNAs were designed in total, each of them targeting the following resistance genes: Ampicillin, Chloramphenicol, Erythromycin, Kanamycin and Spectinomycin. Each gRNA is constituted by a common sequence of the structure DR + 24 bp-spacer + DR + L3S2P21 terminator. The spacer sequence is followed and preceded by a DR since Cas12 cuts the pre-crRNA 4 nucleotides upstream of the hairpin structures formed by the DR. This is important considering that Cas12a can process its own gRNAs (CRISPR RNAs) because of the dual RNase/DNase activity of Cas12a. | ||
| + | |||
| + | We used the <html><a href="https://sfvideo.blob.core.windows.net/sitefinity/docs/default-source/user-guide-manual/dnasealert-detection-manual.pdf?sfvrsn=34133407_8" target="_blank" onmouseover="this.style.color='#ff724a';" onmouseout="this.style.color='#ff9515';" style="color: #ff9515; text-decoration: none; margin-right:-15px;">Substrate Nuclease Detection System</a></html> to perform detection and subsequent fluorescent measurements with the Plate Reader. To give a glimpse of the results we obtained, we present the analysis of a detection graph from the Chloramphenicol gRNA efficient construct. The fluorescence results for <html><a href="https://parts.igem.org/Part:BBa_K2927005" target="_blank" onmouseover="this.style.color='#ff724a';" onmouseout="this.style.color='#ff9515';" style="color: #ff9515; text-decoration: none;">BBa_K2927005</a></html> + <html><a href="https://parts.igem.org/Part:BBa_K3791021" target="_blank" onmouseover="this.style.color='#ff724a';" onmouseout="this.style.color='#ff9515';" style="color: #ff9515; text-decoration: none;">BBa_K3791021</a></html> (Cas12a + gRNA for Chloramphenicol resistant gene) in Fig. 6 range between 26031 and 21607 RFU and are coherent since the one with sample needs to be higher due to the gRNA - sample sequence match. | ||
| + | The decrease in fluorescence signal is only 16.99% over 2 -hour time, which indicates a maintained stability of the signal. The biological significant negative control (without sample) produces a signal which is 38.1% lower in RFU than the one given by the biosensor with its corresponding sample. It ranges between 16118 and 14461 RFU. This is attributed to the nuclease activity released from the lysis process. In both cases, the fluorescence is much higher than in the blank (negative control with water), a fact that is consistent. These results confirm our engineered biological system serves as a biosensor and accomplish the purpose for which it was created. | ||
| + | |||
| + | [[Image:T--UPF_Barcelona--Description_graph_PoC_2and9_white.png|600px|thumb|center|'''Fig 6. Fluorescence measurements for gRNA for Chloramphenicol resistant gene+LbCas12a.''' | ||
| + | ]] | ||
<!-- --> | <!-- --> | ||
| − | |||
<partinfo>BBa_K2927005 SequenceAndFeatures</partinfo> | <partinfo>BBa_K2927005 SequenceAndFeatures</partinfo> | ||
| Line 48: | Line 76: | ||
<partinfo>BBa_K2927005 parameters</partinfo> | <partinfo>BBa_K2927005 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | |||
| + | <h2> <b> Contribution of Squirrel-CHN-II 2024 </b> </h2> | ||
| + | ===New Hair-pin reporter with enhanced trans-cleavage activity of Cas12a.=== | ||
| + | The ssDNA is an important reporter system in Cas12 <i>trans</i>-cleavage experiments, which affects the detection efficiency of the system. ssDNA is currently traditionally sequenced as 5′-(6-FAM)-TTATT-(BHQ1)-3′. During our review of the literature, we found that hairpin DNA with a stem-loop structure can improve the efficiency of Cas12 trans-cleavage (Fig. 1)[1]. | ||
| + | <html> | ||
| + | <head> | ||
| + | <meta charset="utf-8"> | ||
| + | |||
| + | </head> | ||
| + | <body> | ||
| + | <div> | ||
| + | <img src="https://static.igem.wiki/teams/5132/parts/fig-4.webp" style="width: 80%;"> | ||
| + | </div> | ||
| + | </body> | ||
| + | </html> | ||
| + | Fig. 1, Characterize the efficiency of different hairpin DNA reporters[1].<br> | ||
| + | <b>A</b>, Three different hairpin DNA reports were tested that have the same stem composition (3GC + 2AT) but different loop lengths and increasing numbers of thymine (from 5 to 30). <b>B</b>, LbCas12a trans nuclease activity on linear ssDNA and hairpin DNA reporters. <br> | ||
| + | |||
| + | Table1. Sequence of ssDNA reporters[1]. | ||
| + | {|class='wikitable' | ||
| + | |'''Name''' | ||
| + | |'''Sequence''' | ||
| + | |- | ||
| + | |Linear ss-DNA | ||
| + | |5'-(6-FAM)-TTATT–(BHQ1)-3' | ||
| + | |- | ||
| + | |Stem-loop #5T | ||
| + | |5'-(6-FAM)-CTCTCATTTTTAGAGAG–(BHQ1)- 3' | ||
| + | |- | ||
| + | |Stem-loop #10T | ||
| + | |5'-(6-FAM)-CTCTCATTTTTTTTTTAGAGAG-(BHQ1)-3' | ||
| + | |- | ||
| + | |Stem-loop #30T | ||
| + | |5'-(6-FAM)-CTCTCATTTTTTTTTTTTTTTTTTTTTTTTTTTTTTAGAGAG-(BHQ1)-3' | ||
| + | |- | ||
| + | |} | ||
| + | <br> | ||
| + | <b>Reference:</b><br> | ||
| + | [1]Marianna Rossetti, Rosa Merlo, Neda Bagheri, Enhancement of CRISPR/Cas12a <i>trans</i>-cleavage activity using hairpin DNA reporters, <i>Nucleic Acids Research</i>, <b>50</b>(14): 8377–83912022 (2022). | ||
| + | |||
| + | ===Effect of ions on LbCas12a’s collateral cleavage activity=== | ||
| + | ====iGEM Thrace 2021==== | ||
| + | LbCas12a’s collateral cleavage activity can be altered in the presence of particular ions. Specifically, Mn2+ ions can activate the crRNA-independent Dnase activity of the enzyme. Mn2+ is not able to trigger LbCas12a-mediated dsDNA cleavage activity. None of the divalent metal ions is able to trigger LbCas12a-mediated dsDNase activity | ||
| + | |||
| + | ====Reference==== | ||
| + | Li, B., Yan, J., Zhang, Y., Li, W., Zeng, C., Zhao, W., Hou, X., Zhang, C., & Dong, Y. (2020). CRISPR-CAS12A possesses unconventional DNase activity that can be inactivated by synthetic oligonucleotides. Molecular Therapy - Nucleic Acids, 19, 1043–1052. https://doi.org/10.1016/j.omtn.2019.12.038 | ||
| + | |||
| + | |||
| + | ===Improvement of LbCas12a=== | ||
| + | ====iGEM NTU-Singapore 2021==== | ||
| + | The improved version of this part is part BBa_K4053001 | ||
| + | |||
| + | An evolved mutant of LbCas12a, LbCas12a-RVRR with mutations at G532R, K538V, Y542R and K595R. | ||
| + | |||
| + | The LbCas12a-RVRR has a more varied PAM, achieved by introducing the mutations at the analogous positions of the RVR and RR variants of LbCas12a. As the trans-cleavage activity of the LbCas12a-RVRR was not characterised, we decided to test its activity to detect point mutation. We detected the same target (BRAF V600E) and we managed to further improve the sensitivity for the same target mutation detection. We made several mutations for WT-LbCas12a plasmid and managed to purify the LbCas12a-RVRR. We also tested it in the FQ Assay and found out that it can better differentiate mutant: wild-type template, hence providing higher sensitivity for snip detection. | ||
| + | |||
| + | ====Results==== | ||
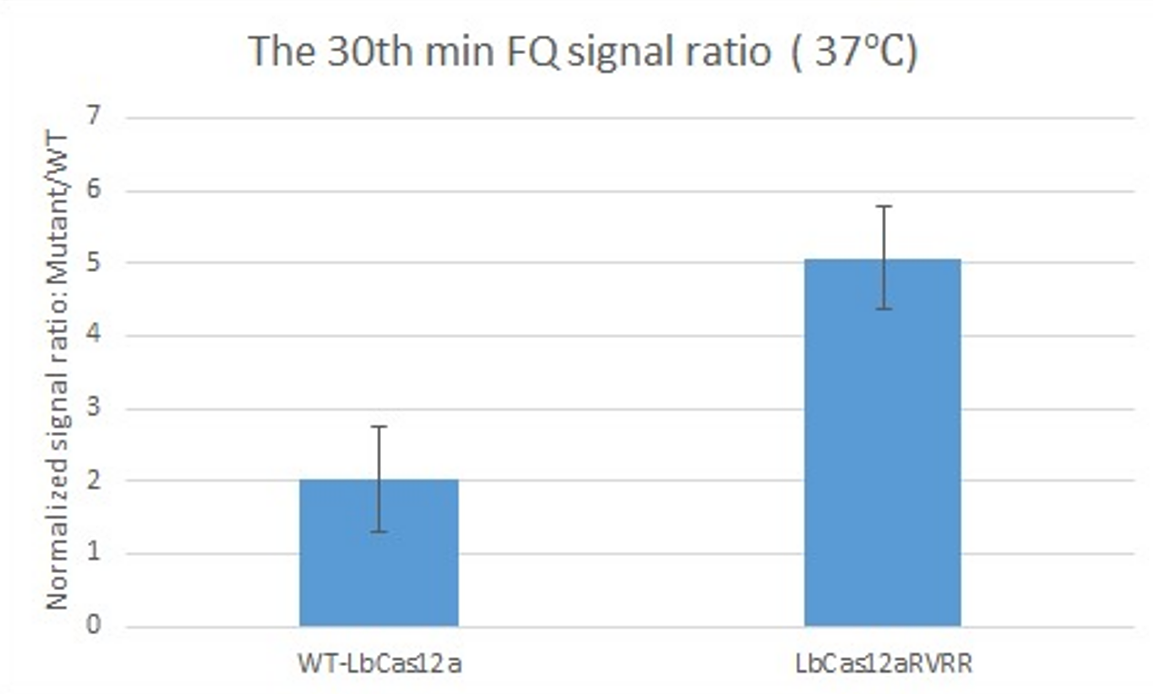
| + | After complexing WT-LbCas12a and LbCas12a-RVRR to the gRNAs, we tested their detection performance against BRAF-V600E by using the FQ (Fluorophore-Quencher) reaction and recorded the fluorescence signal. | ||
| + | |||
| + | [[File:T--NTU-Singapore-LbCas12aRVRR.png|center|500px|]] | ||
| + | |||
| + | The figure above shows the signal of ratio:mutant (BRAF-V600:BRAF-E600) for WT-LbCas12a (left) and LbCas12a-RVRR (right) recorded at the 30th minute in 37°C in the FQ reaction. | ||
| + | |||
| + | The higher signal from LbCas12a-RVRR shows that LbCas12a-RVRR can differentiate BRAF-V600E better than WT-LbCas12a. Therefore, our experiment has proven that LbCas12a-RVRR has a higher sensitivity towards mutation in the BRAF gene. | ||
| + | |||
| + | ====References==== | ||
| + | Tóth, E., Varga, É., Kulcsár, P. I., Kocsis-Jutka, V., Krausz, S. L., Nyeste, A., Welker, Z., Huszár, K., Ligeti, Z., Tálas, A., & Welker, E. (2020). Improved LBCAS12A variants with altered pam specificities further broaden the genome targeting range of Cas12a nucleases. Nucleic Acids Research, 48(7), 3722–3733. https://doi.org/10.1093/nar/gkaa110 | ||
Latest revision as of 08:15, 27 September 2024
LbCas12a
CRISPR Cas12a system is one of the bacterial adaptive immune systems. Cas12a protein is the RNA-guided enzyme that binds and cut DNA. When Cas12a protein bind with the specific crRNA, it will be activated. After Cas12a protein is activated, it will cut the target DNA as well as non-specific single-strand DNA (ssDNA), this certain function is used in our project. We designed six crRNA which is derived from the African swine fever virus (ASFV) to detect the DNA of the virus. This sequence is just a part of DNA sequence of the vp72 membrane protein of ASFV.
Reference
Janice S. Chen, Enbo Ma, Lucas B. Harrington, Maria Da Costa, Xinran Tian, Joel M. Palefsky, Jennifer A. Doudna, Chen et al., Science 360, 436–439 (2018)
Experiment Results
- Introduction
In our project, we combined three parts of biological reactions to detect ASFV specific sequence in samples. The first one is LbCas12a-crRNA system, which can specifically recognize ASFV specific double stranded DNA (dsDNA) sequence on P72 gene. The secondary part is the trans-activation of LbCas12a-crRNA system. When LbCas12a-crRNA system binds to ASFV specific dsDNA sequences, LbCas12a-crRNA system will cleave dsDNA and further degrade non-specific single stranded DNA (ssDNA). To detect the degradation of ssDNA in ASFV-activated LbCas12a-crRNA system, we will use the PicoGreen fluoresce dye to monitor the undegraded ssDNA, which is the third part. To transfer reaction from part I/II/III to detection, we plane to conjugate ssDNA on magnetic beads. The ssDNA conjugated magnetic beads will be easily captured and transfer by electromagnetic force. In the following result section, we will show our progress through experiments that supported our project design.
- Our targets
Steps to establish CRISPR-LbCas12a system
Expression of LbCas12a protein: We transformed pHMT-LbCas12a into E.coli BL21, and then added 0.2 mM IPTG to induce protein expression (see notebook for details). The result showed that the LbCas12a protein expression in soluble fraction was induced by IPTG, and increased as time goes on (Figure 1). The predicted protein size of LbCas12a with MBP and His-tag is about 180 kDa, which is close to the induced protein indicted by red arrow in figure 1. We also examined the insoluble fraction of IPTG induced BL21 by SDS-PAGE, and confirmed that most LbCas12a protein was soluble (Figure 2).
Pre-test of LbCas12a protein purification:
After confirming the induction of LbCas12a protein expression, we purify LbCas12a protein by Ni2+-magnetic beads to pull down the His-tag on LbCas12a protein from soluble fraction. The elution of LbCas12a protein from Ni2+-magnetic beads by excess imidazole or TEV enzyme digestion further confirmed that the Ni2+-magnetic beads purification is clear and easy to reverse (Figure 3).
According to the experiments above, we can express and purified LbCas12a proteins for further application. Therefore we scale up the expression and purification of LbCas12a protein.
Large scale protein purification:
We use immobilized metal affinity chromatography (IMAC) to purify LbCas12a protein from soluble fraction of BL21 by Ni2+ chelating sepharose column. We then elute LbCas12a protein from Ni2+ column by imidazole and subjected into FPLC separation. The absorption peak at 27-30 fractions was indicated by red arrow in figure 4.
To further confirm that the absorption peak, we performed SDS-PAGE and Coomassie blue staining to fractions 27-30, showing that the absorption peak is indeed LbCas12a protein.
UPF Barcelona ARIA Team experimental application
Usage, Biology and Characterization
ARIA’s biosensors are based on two main elements: the endonuclease enzyme LbCas12a from Lachnospiraceae Bacterium, and the crRNAs designed specifically to target antibiotic resistance (AR) sequences. Once the two elements bind and the target is detected, a fluorescent signal is activated due to LbCas12a cis cleavage activity. However, these elements have been implemented on living E.Coli (BL21), and become active after lysing the cells and being put in contact with AR sequences. This has given rise to a new innovative approach: the in vivo – in vitro detection.
To do so, the original plasmid employed for cloning Cas12 was taken from (Addgene), and it was optimized by deleting purification sites and unuseful sequences codifying for MBP (Maltose Binding Protein). Cas12 is expressed under the T7 promoter so that it is inducible with IPTG, and it has ampicillin resistance to allow for its selection.
5 gRNAs were designed in total, each of them targeting the following resistance genes: Ampicillin, Chloramphenicol, Erythromycin, Kanamycin and Spectinomycin. Each gRNA is constituted by a common sequence of the structure DR + 24 bp-spacer + DR + L3S2P21 terminator. The spacer sequence is followed and preceded by a DR since Cas12 cuts the pre-crRNA 4 nucleotides upstream of the hairpin structures formed by the DR. This is important considering that Cas12a can process its own gRNAs (CRISPR RNAs) because of the dual RNase/DNase activity of Cas12a.
We used the Substrate Nuclease Detection System to perform detection and subsequent fluorescent measurements with the Plate Reader. To give a glimpse of the results we obtained, we present the analysis of a detection graph from the Chloramphenicol gRNA efficient construct. The fluorescence results for BBa_K2927005 + BBa_K3791021 (Cas12a + gRNA for Chloramphenicol resistant gene) in Fig. 6 range between 26031 and 21607 RFU and are coherent since the one with sample needs to be higher due to the gRNA - sample sequence match. The decrease in fluorescence signal is only 16.99% over 2 -hour time, which indicates a maintained stability of the signal. The biological significant negative control (without sample) produces a signal which is 38.1% lower in RFU than the one given by the biosensor with its corresponding sample. It ranges between 16118 and 14461 RFU. This is attributed to the nuclease activity released from the lysis process. In both cases, the fluorescence is much higher than in the blank (negative control with water), a fact that is consistent. These results confirm our engineered biological system serves as a biosensor and accomplish the purpose for which it was created.
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1371
Illegal BglII site found at 2108 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 3022
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 1435
Contribution of Squirrel-CHN-II 2024
New Hair-pin reporter with enhanced trans-cleavage activity of Cas12a.
The ssDNA is an important reporter system in Cas12 trans-cleavage experiments, which affects the detection efficiency of the system. ssDNA is currently traditionally sequenced as 5′-(6-FAM)-TTATT-(BHQ1)-3′. During our review of the literature, we found that hairpin DNA with a stem-loop structure can improve the efficiency of Cas12 trans-cleavage (Fig. 1)[1].

A, Three different hairpin DNA reports were tested that have the same stem composition (3GC + 2AT) but different loop lengths and increasing numbers of thymine (from 5 to 30). B, LbCas12a trans nuclease activity on linear ssDNA and hairpin DNA reporters.
Table1. Sequence of ssDNA reporters[1].
| Name | Sequence |
| Linear ss-DNA | 5'-(6-FAM)-TTATT–(BHQ1)-3' |
| Stem-loop #5T | 5'-(6-FAM)-CTCTCATTTTTAGAGAG–(BHQ1)- 3' |
| Stem-loop #10T | 5'-(6-FAM)-CTCTCATTTTTTTTTTAGAGAG-(BHQ1)-3' |
| Stem-loop #30T | 5'-(6-FAM)-CTCTCATTTTTTTTTTTTTTTTTTTTTTTTTTTTTTAGAGAG-(BHQ1)-3' |
Reference:
[1]Marianna Rossetti, Rosa Merlo, Neda Bagheri, Enhancement of CRISPR/Cas12a trans-cleavage activity using hairpin DNA reporters, Nucleic Acids Research, 50(14): 8377–83912022 (2022).
Effect of ions on LbCas12a’s collateral cleavage activity
iGEM Thrace 2021
LbCas12a’s collateral cleavage activity can be altered in the presence of particular ions. Specifically, Mn2+ ions can activate the crRNA-independent Dnase activity of the enzyme. Mn2+ is not able to trigger LbCas12a-mediated dsDNA cleavage activity. None of the divalent metal ions is able to trigger LbCas12a-mediated dsDNase activity
Reference
Li, B., Yan, J., Zhang, Y., Li, W., Zeng, C., Zhao, W., Hou, X., Zhang, C., & Dong, Y. (2020). CRISPR-CAS12A possesses unconventional DNase activity that can be inactivated by synthetic oligonucleotides. Molecular Therapy - Nucleic Acids, 19, 1043–1052. https://doi.org/10.1016/j.omtn.2019.12.038
Improvement of LbCas12a
iGEM NTU-Singapore 2021
The improved version of this part is part BBa_K4053001
An evolved mutant of LbCas12a, LbCas12a-RVRR with mutations at G532R, K538V, Y542R and K595R.
The LbCas12a-RVRR has a more varied PAM, achieved by introducing the mutations at the analogous positions of the RVR and RR variants of LbCas12a. As the trans-cleavage activity of the LbCas12a-RVRR was not characterised, we decided to test its activity to detect point mutation. We detected the same target (BRAF V600E) and we managed to further improve the sensitivity for the same target mutation detection. We made several mutations for WT-LbCas12a plasmid and managed to purify the LbCas12a-RVRR. We also tested it in the FQ Assay and found out that it can better differentiate mutant: wild-type template, hence providing higher sensitivity for snip detection.
Results
After complexing WT-LbCas12a and LbCas12a-RVRR to the gRNAs, we tested their detection performance against BRAF-V600E by using the FQ (Fluorophore-Quencher) reaction and recorded the fluorescence signal.
The figure above shows the signal of ratio:mutant (BRAF-V600:BRAF-E600) for WT-LbCas12a (left) and LbCas12a-RVRR (right) recorded at the 30th minute in 37°C in the FQ reaction.
The higher signal from LbCas12a-RVRR shows that LbCas12a-RVRR can differentiate BRAF-V600E better than WT-LbCas12a. Therefore, our experiment has proven that LbCas12a-RVRR has a higher sensitivity towards mutation in the BRAF gene.
References
Tóth, E., Varga, É., Kulcsár, P. I., Kocsis-Jutka, V., Krausz, S. L., Nyeste, A., Welker, Z., Huszár, K., Ligeti, Z., Tálas, A., & Welker, E. (2020). Improved LBCAS12A variants with altered pam specificities further broaden the genome targeting range of Cas12a nucleases. Nucleic Acids Research, 48(7), 3722–3733. https://doi.org/10.1093/nar/gkaa110