Difference between revisions of "Part:BBa K2460009"
Zongyeqing (Talk | contribs) (→Experimental Setup) |
(→Characterization by GENAS_China 2019) |
||
| Line 8: | Line 8: | ||
=Characterization by GENAS_China 2019= | =Characterization by GENAS_China 2019= | ||
*'''Group:''' GENAS_China 2019 | *'''Group:''' GENAS_China 2019 | ||
| − | We | + | We used [[Part:BBa_K2460009|BBa_K2460009]] and this part (placed in the same direction) as functional elements in [[Part:BBa_K3254005|BBa_K3254005]], and the DNA between [[Part:BBa_K2460008|BBa_K2460008]] and [[Part:BBa_K2460009|BBa_K2460009]] was excised via recombination by TG1 integrase as expected, see below. |
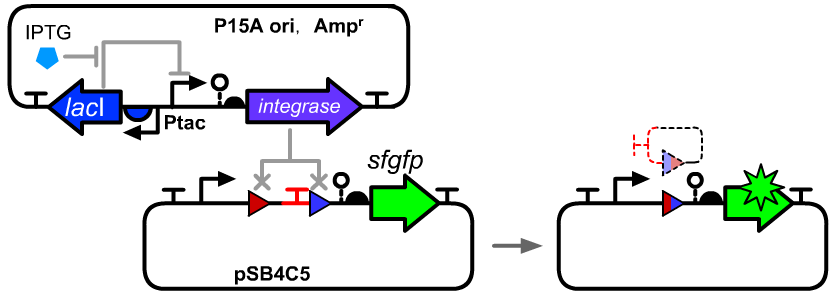
==Genetic Design== | ==Genetic Design== | ||
| − | The composition and principle of the experimental system | + | The composition and principle of the experimental system were indicated below. More details can be seen on the pages of [[Part:BBa_K3254013|BBa_K3254013]] and [[Part:BBa_K3254005|BBa_K3254005]]. |
[[File:T--GENAS_China--excision_with_backbone.PNG|200px|thumb|left| ]] | [[File:T--GENAS_China--excision_with_backbone.PNG|200px|thumb|left| ]] | ||
| Line 17: | Line 17: | ||
==Experimental Setup== | ==Experimental Setup== | ||
The reporter plasmid contained this part and [[Part:BBa_K2460009|BBa_K2460009]] were co-transferred into E.coli DH5α host with 6 integrase generator plasmids. | The reporter plasmid contained this part and [[Part:BBa_K2460009|BBa_K2460009]] were co-transferred into E.coli DH5α host with 6 integrase generator plasmids. | ||
| − | Then single colonies were inoculated into M9 supplemented medium for overnight growth. Then, the cell cultures were diluted 1000-fold with M9 supplemented medium with 500 μM IPTG inducer and growth for another 20 hours. All incubations were carried out using a Digital Thermostatic Shaker maintained at 37 °C and 1000 rpm, using flat-bottom 96-well plates sealed with sealing film. Finally, | + | Then single colonies were inoculated into M9 supplemented medium for overnight growth. Then, the cell cultures were diluted 1000-fold with M9 supplemented medium with 500 μM IPTG inducer and growth for another 20 hours. All incubations were carried out using a Digital Thermostatic Shaker maintained at 37 °C and 1000 rpm, using flat-bottom 96-well plates sealed with sealing film. Finally, the cultures were sampled for genotype PCR testing. |
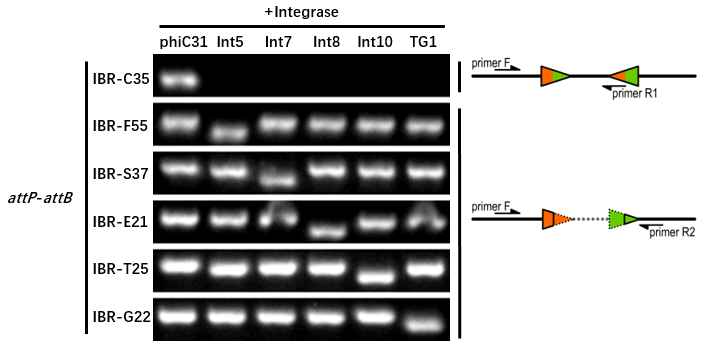
The principle of genotype identification was shown on the right of results image. | The principle of genotype identification was shown on the right of results image. | ||
*M9 medium (supplemented): 6.8 g/L Na<sub>2</sub>HPO<sub>4</sub>, 3 g/L KH<sub>2</sub>PO<sub>4</sub>, 0.5 g/L NaCl, 1 g/L NH<sub>4</sub>Cl, 0.34 g/L thiamine, 0.2% casamino acids, 0.4% glucose, 2mM MgSO<sub>4</sub>, and 100 μM CaCl<sub>2</sub>. | *M9 medium (supplemented): 6.8 g/L Na<sub>2</sub>HPO<sub>4</sub>, 3 g/L KH<sub>2</sub>PO<sub>4</sub>, 0.5 g/L NaCl, 1 g/L NH<sub>4</sub>Cl, 0.34 g/L thiamine, 0.2% casamino acids, 0.4% glucose, 2mM MgSO<sub>4</sub>, and 100 μM CaCl<sub>2</sub>. | ||
| Line 23: | Line 23: | ||
==Results== | ==Results== | ||
IBR-G22 was the plasmid with TG1 attB and attP sites. | IBR-G22 was the plasmid with TG1 attB and attP sites. | ||
| − | The | + | The results indicated that the TG1 att sites could only be recombined by TG1 integrase. |
| − | The sequence after recombination (attL or attR) | + | The sequence after recombination (attL or attR) was GATCAGCTCCGCGGGCAAGACCGTGCTCTTACCCAGTTGGGCGGGA. |
[[File:T--GENAS_China--orthogonality_test.png]] | [[File:T--GENAS_China--orthogonality_test.png]] | ||
| Line 33: | Line 33: | ||
<!-- --> | <!-- --> | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
| − | <partinfo>BBa_K2460009 | + | <partinfo>BBa_K2460009 Sequence And Features</partinfo> |
Latest revision as of 16:14, 21 October 2019
TG1 attP
This is a recombination site attP for TG1 integrase (BBa_K2460007). When used with phiBT1 attB (BBa_K2460008) and TG1 integrase, it can flip a DNA sequence flanked by attB and attP sites, as well as integrating a circular DNA with a attB/P site into a linear/circular DNA with a attP/B site. For more information about kinetic characterization and the orthogonality between the integrase and some other serine recombinase, go to TG1 integrase.
Characterization by GENAS_China 2019
- Group: GENAS_China 2019
We used BBa_K2460009 and this part (placed in the same direction) as functional elements in BBa_K3254005, and the DNA between BBa_K2460008 and BBa_K2460009 was excised via recombination by TG1 integrase as expected, see below.
Genetic Design
The composition and principle of the experimental system were indicated below. More details can be seen on the pages of BBa_K3254013 and BBa_K3254005.
Experimental Setup
The reporter plasmid contained this part and BBa_K2460009 were co-transferred into E.coli DH5α host with 6 integrase generator plasmids. Then single colonies were inoculated into M9 supplemented medium for overnight growth. Then, the cell cultures were diluted 1000-fold with M9 supplemented medium with 500 μM IPTG inducer and growth for another 20 hours. All incubations were carried out using a Digital Thermostatic Shaker maintained at 37 °C and 1000 rpm, using flat-bottom 96-well plates sealed with sealing film. Finally, the cultures were sampled for genotype PCR testing. The principle of genotype identification was shown on the right of results image.
- M9 medium (supplemented): 6.8 g/L Na2HPO4, 3 g/L KH2PO4, 0.5 g/L NaCl, 1 g/L NH4Cl, 0.34 g/L thiamine, 0.2% casamino acids, 0.4% glucose, 2mM MgSO4, and 100 μM CaCl2.
Results
IBR-G22 was the plasmid with TG1 attB and attP sites. The results indicated that the TG1 att sites could only be recombined by TG1 integrase. The sequence after recombination (attL or attR) was GATCAGCTCCGCGGGCAAGACCGTGCTCTTACCCAGTTGGGCGGGA.
Sequence and Features BBa_K2460009 Sequence And Features Not understood