Difference between revisions of "Part:BBa K3260025"
Maryeliz46 (Talk | contribs) |
Maryeliz46 (Talk | contribs) |
||
| (12 intermediate revisions by the same user not shown) | |||
| Line 10: | Line 10: | ||
It was designed to be a basic hydrophobic domain to include in a variety of iterations of cellulose binding, hydrophobic proteins. Used in the composite part BBa_K3260031, where it was fused to a dCBD using a flex linker, and was the basis of another basic part, BBa_K3260035. | It was designed to be a basic hydrophobic domain to include in a variety of iterations of cellulose binding, hydrophobic proteins. Used in the composite part BBa_K3260031, where it was fused to a dCBD using a flex linker, and was the basis of another basic part, BBa_K3260035. | ||
| − | <p><i>Implementing Protein-Based Paper Microfluidics using | + | ===<p><i>Implementing Protein-Based Paper Microfluidics using BBa_K3260025</i></p>=== |
<p>BBa_K3260031 Single Layer Print:</p><br> | <p>BBa_K3260031 Single Layer Print:</p><br> | ||
| − | + | https://2019.igem.org/wiki/images/2/22/T--BrownStanfordPrinctn--test_one.mov | |
| − | + | <p>The video linked above shows one of our tests with printing with protein lysate. The lysate that we loaded into the black ink cartridge contained BBa_K3260031(BBa_K1321340+ K3260025). Directly below is a labelling of this video with channel widths for each component in our testing template.</p> | |
| − | + | ||
| − | |||
| − | + | https://2019.igem.org/wiki/images/a/a6/T--BrownStanfordPrinctn--video_one_labels.png | |
<p>The blue regions are plain printer ink, while to the white sections in between the blue and the black outline of the channels are printed protein lysate. We can see on the range of straight, separated channels that the water efficiently wicked up the 250um wide channel and the 200um channel. On the largest “flower” template, the water flowed up the 800um and 900um channels. We also see a successful serendipitous negative control of our device once the water flows past the end of one of the 900um channels and starts penetrating the paper printed with blue ink. This provides a nice contrast with the channel flow and illustrates that while the resolution of the channels isn’t extremely sharp, ===the hydrophobic protein (BBa_K3260025) is certainly performing its function.=== On the smaller scale flower channels, the water efficiently travels down the 100 - 500 um channels, which is indicative of its functionality on the PDMS-scale resolution. | <p>The blue regions are plain printer ink, while to the white sections in between the blue and the black outline of the channels are printed protein lysate. We can see on the range of straight, separated channels that the water efficiently wicked up the 250um wide channel and the 200um channel. On the largest “flower” template, the water flowed up the 800um and 900um channels. We also see a successful serendipitous negative control of our device once the water flows past the end of one of the 900um channels and starts penetrating the paper printed with blue ink. This provides a nice contrast with the channel flow and illustrates that while the resolution of the channels isn’t extremely sharp, ===the hydrophobic protein (BBa_K3260025) is certainly performing its function.=== On the smaller scale flower channels, the water efficiently travels down the 100 - 500 um channels, which is indicative of its functionality on the PDMS-scale resolution. | ||
| Line 24: | Line 22: | ||
<p>BBa_K3260031 Double Layer and Negative Control Side by Side:</p> | <p>BBa_K3260031 Double Layer and Negative Control Side by Side:</p> | ||
| − | |||
| − | |||
| − | + | https://2019.igem.org/wiki/images/9/9f/T--BrownStanfordPrinctn--test_two.mov | |
| − | <p> | + | <p>The video linked above is another printing lysate test. This test print was part negative control (single layer, but underprinted with normal ink) and part double layer printed BBa_K3260031, which is a dCBD fused to a single hydrophobic domain. Below is a labelling of the video with channel width for each component in our testing template.</p> |
| − | We were able to create functional microfluidic channels down to 100um in width, which is extremely significant because this falls comfortably within the range of high resolution PDMS device channels (which have widths of 30 nm to 500 um [2, 3]). Even though our channels did experience a significant degree of bleeding, the functionality of their wicking ability even down to 100um widths speaks to the effectiveness of the hydrophobic domain used in this fusion, BBa_K3260025.</p> | + | https://2019.igem.org/wiki/images/f/fa/T--BrownStanfordPrinctn--video_two_labels.png |
| + | |||
| + | <p> On this test we have a firmer grasp on the resolution that is achieved with this printing method, and it is shown that some bleeding into the hydrophobic region of the device design occurs. However, this is sharply contrasted with the complete lack of resolution and function in the negative control, that was single layer printed with our hydrophobic protein but printed on the opposite side with plain blue ink, which undercuts the functionality of the protein. We can see that the water effectively travels up the channels of widths 200 um, 250 um, 800 um, and 900 um. <br><br> | ||
| + | |||
| + | We were able to create functional microfluidic channels down to 100um in width, which is extremely significant because this falls comfortably within the range of high resolution PDMS device channels (which have widths of 30 nm to 500 um [2, 3]). Even though our channels did experience a significant degree of bleeding, the <b> functionality of their wicking ability even down to 100um widths speaks to the effectiveness of the hydrophobic domain used in this fusion, BBa_K3260025.</b></p> | ||
<br><p>[1] Liu, Ning, et al. “Direct Spraying Method for Fabrication of Paper-Based Microfluidic Devices.” Semantic Scholar , Nanyang Technological University , Sept. 2017.</p> | <br><p>[1] Liu, Ning, et al. “Direct Spraying Method for Fabrication of Paper-Based Microfluidic Devices.” Semantic Scholar , Nanyang Technological University , Sept. 2017.</p> | ||
Latest revision as of 15:12, 21 October 2019
Hydrophobic Domain
Radek Hydrophobic Domain
A hydrophobic domain comprised of 12 phenylalanines, with a variety of codons used to increase stability as a gblock. Proposed by Dr. Radoslaw Piast from the University of Warsaw, who briefly was a visiting scientist at the NASA Ames Research Center.
Usage and Biology
It was designed to be a basic hydrophobic domain to include in a variety of iterations of cellulose binding, hydrophobic proteins. Used in the composite part BBa_K3260031, where it was fused to a dCBD using a flex linker, and was the basis of another basic part, BBa_K3260035.
Implementing Protein-Based Paper Microfluidics using BBa_K3260025
BBa_K3260031 Single Layer Print:
https://2019.igem.org/wiki/images/2/22/T--BrownStanfordPrinctn--test_one.mov
The video linked above shows one of our tests with printing with protein lysate. The lysate that we loaded into the black ink cartridge contained BBa_K3260031(BBa_K1321340+ K3260025). Directly below is a labelling of this video with channel widths for each component in our testing template.
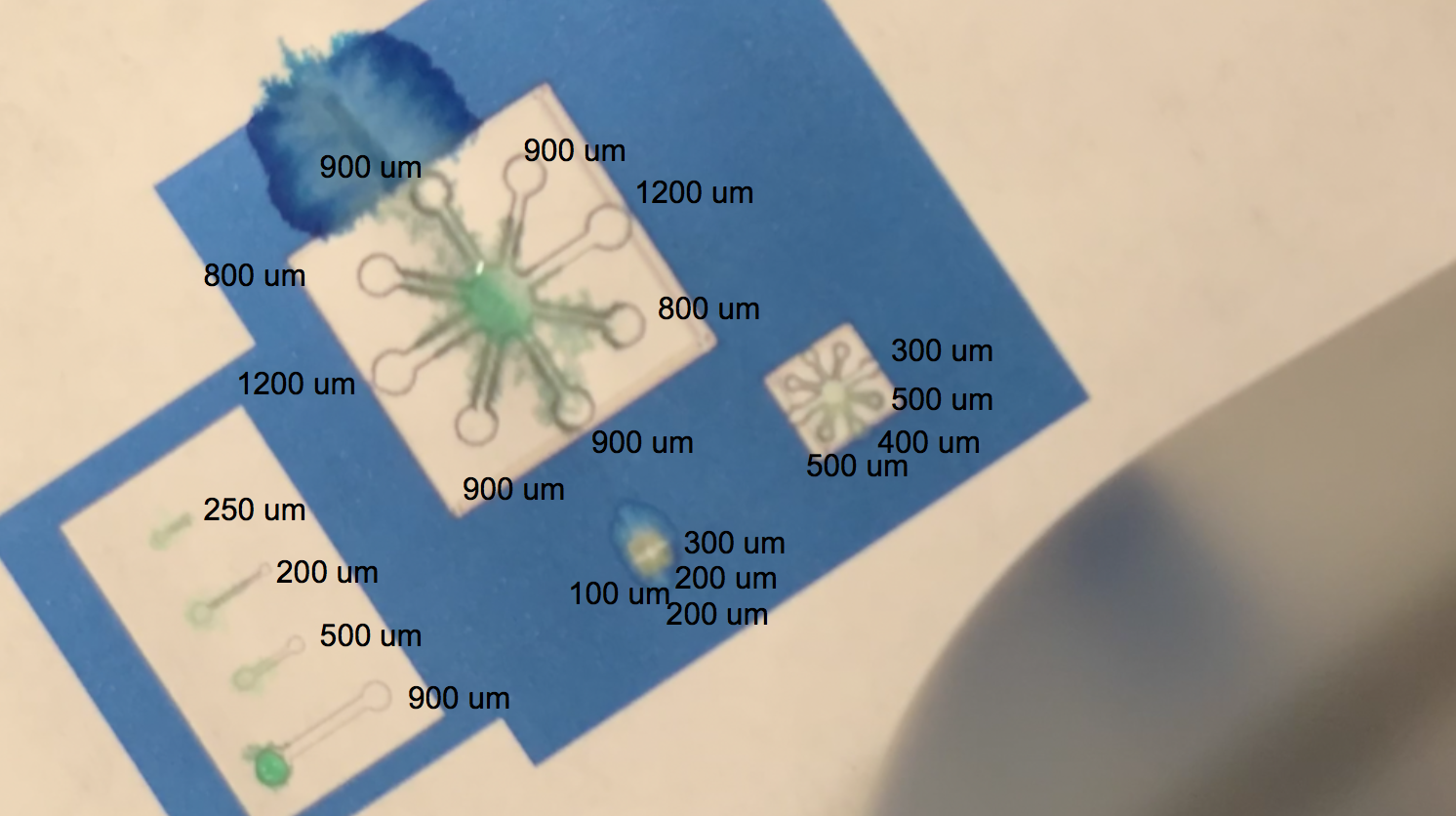
The blue regions are plain printer ink, while to the white sections in between the blue and the black outline of the channels are printed protein lysate. We can see on the range of straight, separated channels that the water efficiently wicked up the 250um wide channel and the 200um channel. On the largest “flower” template, the water flowed up the 800um and 900um channels. We also see a successful serendipitous negative control of our device once the water flows past the end of one of the 900um channels and starts penetrating the paper printed with blue ink. This provides a nice contrast with the channel flow and illustrates that while the resolution of the channels isn’t extremely sharp, ===the hydrophobic protein (BBa_K3260025) is certainly performing its function.=== On the smaller scale flower channels, the water efficiently travels down the 100 - 500 um channels, which is indicative of its functionality on the PDMS-scale resolution.
BBa_K3260031 Double Layer and Negative Control Side by Side:
https://2019.igem.org/wiki/images/9/9f/T--BrownStanfordPrinctn--test_two.mov
The video linked above is another printing lysate test. This test print was part negative control (single layer, but underprinted with normal ink) and part double layer printed BBa_K3260031, which is a dCBD fused to a single hydrophobic domain. Below is a labelling of the video with channel width for each component in our testing template.

On this test we have a firmer grasp on the resolution that is achieved with this printing method, and it is shown that some bleeding into the hydrophobic region of the device design occurs. However, this is sharply contrasted with the complete lack of resolution and function in the negative control, that was single layer printed with our hydrophobic protein but printed on the opposite side with plain blue ink, which undercuts the functionality of the protein. We can see that the water effectively travels up the channels of widths 200 um, 250 um, 800 um, and 900 um.
We were able to create functional microfluidic channels down to 100um in width, which is extremely significant because this falls comfortably within the range of high resolution PDMS device channels (which have widths of 30 nm to 500 um [2, 3]). Even though our channels did experience a significant degree of bleeding, the functionality of their wicking ability even down to 100um widths speaks to the effectiveness of the hydrophobic domain used in this fusion, BBa_K3260025.
[1] Liu, Ning, et al. “Direct Spraying Method for Fabrication of Paper-Based Microfluidic Devices.” Semantic Scholar , Nanyang Technological University , Sept. 2017.
[2] Abidin, Ummikalsom, et al. “Replication and Leakage Test of Polydimethylsiloxane (PDMS) Microfluidics Channel.” AIP Publishing, AIP Publishing LLC, 25 Jan. 2019, aip.scitation.org/doi/abs/10.1063/1.5086611.
[3] Strong, E. Brandon, et al. “Fabrication of Miniaturized Paper-Based Microfluidic Devices (MicroPADs).” Nature News, Nature Publishing Group, 9 Jan. 2019, www.nature.com/articles/s41598-018-37029-0.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
