Difference between revisions of "Part:BBa K3081002"
| (2 intermediate revisions by the same user not shown) | |||
| Line 5: | Line 5: | ||
This part consists of araC/araBAD promoter and sfGFP fused to the C terminus of dCas9. Expression level of dCas9 induced by arabinose is tested by fluorescence of fused dCas9 linker sfGFP. | This part consists of araC/araBAD promoter and sfGFP fused to the C terminus of dCas9. Expression level of dCas9 induced by arabinose is tested by fluorescence of fused dCas9 linker sfGFP. | ||
| − | Promoter pBAD is induced with inexpensive and non-toxic monosaccharide L-arabinose and in bacterial strains deficient in arabinose catabolism. This promoter is tightly regulated and can reach moderately high levels of downstream gene expression.In order to obtain a dynamic range and proper concentration to induce the expression of dCas9, we use | + | Promoter pBAD is induced with inexpensive and non-toxic monosaccharide L-arabinose and in bacterial strains deficient in arabinose catabolism. This promoter is tightly regulated and can reach moderately high levels of downstream gene expression.In order to obtain a dynamic range and proper concentration to induce the expression of dCas9, we use cytometry to characterize the pBAD/araC promoter on respectively medium copy number pSB3C5 and low copy number pSB4A5 in TOP10 and found the fluorescense is increased as the concentration of arabinose increases (Figure 1).After we changed the downstream gene "sfGFP" with "sfGFP fused dCas9" on pSB3C5, it was observed that the expression of dCas9 from pBAD promoter can be modulated over a wide range of arabinose concentrations (Figure 2). |
<center> | <center> | ||
| Line 20: | Line 20: | ||
</center> | </center> | ||
| + | In order to alleviate the burden caused by dCas9,we add a degradation tag "ssrA" to the end of sfGFP. Then we found that the protein with ssrA was expressed at a lower level than the control protein, with the same concentration of inducer. | ||
| + | |||
| + | <center> | ||
| + | https://2019.igem.org/wiki/images/7/78/T--Peking--dls_ssrA.png | ||
| + | </center> | ||
| + | <center> | ||
| + | Figure 3: Comparison between dCas9 and dCas9-ssrA system by expression level (on a low copy number plasmid pSB4A5) | ||
| + | </center> | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Latest revision as of 20:28, 21 October 2019
pBAD-dCas9-linker-sfGFP
This part consists of araC/araBAD promoter and sfGFP fused to the C terminus of dCas9. Expression level of dCas9 induced by arabinose is tested by fluorescence of fused dCas9 linker sfGFP.
Promoter pBAD is induced with inexpensive and non-toxic monosaccharide L-arabinose and in bacterial strains deficient in arabinose catabolism. This promoter is tightly regulated and can reach moderately high levels of downstream gene expression.In order to obtain a dynamic range and proper concentration to induce the expression of dCas9, we use cytometry to characterize the pBAD/araC promoter on respectively medium copy number pSB3C5 and low copy number pSB4A5 in TOP10 and found the fluorescense is increased as the concentration of arabinose increases (Figure 1).After we changed the downstream gene "sfGFP" with "sfGFP fused dCas9" on pSB3C5, it was observed that the expression of dCas9 from pBAD promoter can be modulated over a wide range of arabinose concentrations (Figure 2).
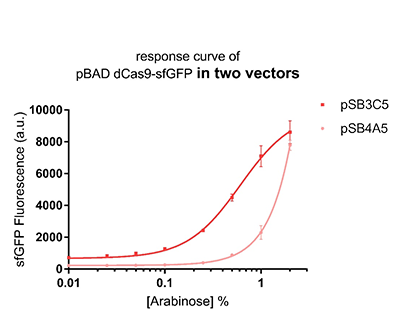
Figure 1: The response curve of pBAD dCas9-sfGFP in two vectors, respectively medium copy number pSB3C5 and low copy number pSB4A5
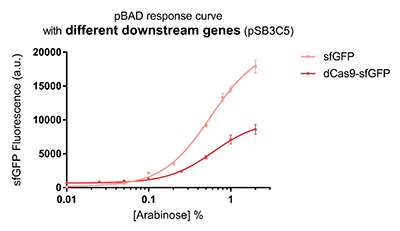
Figure 2: pBAD response curve with different downstream genes
In order to alleviate the burden caused by dCas9,we add a degradation tag "ssrA" to the end of sfGFP. Then we found that the protein with ssrA was expressed at a lower level than the control protein, with the same concentration of inducer.

Figure 3: Comparison between dCas9 and dCas9-ssrA system by expression level (on a low copy number plasmid pSB4A5)
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1205
Illegal NheI site found at 2321 - 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 1144
Illegal BamHI site found at 4600 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 979
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 961
Illegal SapI.rc site found at 5369
