Difference between revisions of "Part:BBa K3100022"
SquarePants (Talk | contribs) |
|||
| (6 intermediate revisions by 2 users not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K3100022 short</partinfo> | <partinfo>BBa_K3100022 short</partinfo> | ||
| − | <strong>Usage and Biology:</strong><br> | + | <h2><strong>Usage and Biology:</strong><br></h2> |
| − | PT7 is the most well-known inducible promoter with high transcriptional strength[1]. T7 RNAP is a single-subunit RNA polymerase that is a strong driver of transcription. It is functionally orthogonal to most hosts, acting only on its cognate promoter, PT7. In order to achieve a larger scale and more accurate regulation range, we have improved T7 promoters with different strength providing more options for the precise regulation. | + | PT7 is the most well-known inducible promoter with high transcriptional strength[1]. T7 RNAP is a single-subunit RNA polymerase that is a strong driver of transcription. It is functionally orthogonal to most hosts, acting only on its cognate promoter, PT7. In order to achieve a larger scale and more accurate regulation range, we have improved T7 promoters (BBa_j64997)with different strength providing more options for the precise regulation. |
| − | < | + | <center>[[File:T--SCUT China--PT7 site.jpeg|500px]]</center> |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | </ | + | |
<p style="text-align:center;">Fig1. T7 promoters with different strength</p> | <p style="text-align:center;">Fig1. T7 promoters with different strength</p> | ||
| − | <strong>Characterization:</strong><br> | + | <h2><strong>Characterization:</strong></h2><br> |
<strong>1. Construction of the T7 Promoter Library</strong><br> | <strong>1. Construction of the T7 Promoter Library</strong><br> | ||
| − | + | A T7 promoter library was constructed through site-saturation mutagenesis, and an efficient screening method based on GFP fluorescence detection was established.<br> | |
The site-saturation mutagenesis DNA fragment pET30a(+)-PT7-RFP vector 1 was obtained using the plasmid pET30a(+)-PT7-RFP as the template and vector 1.F (the sequence containing the mutation in promoter)and vector 1.R as the primers. At the same time, vector 2.F and vector 2.R as the primers to obtain pET30a(+)-PT7-RFP vector 1 by PCR. And then construct pET30a(+)-PT7 mutagenesis -RFP with pET30a(+)-PT7-RFP vector 1 and vector 2 by Gibson Assembly. | The site-saturation mutagenesis DNA fragment pET30a(+)-PT7-RFP vector 1 was obtained using the plasmid pET30a(+)-PT7-RFP as the template and vector 1.F (the sequence containing the mutation in promoter)and vector 1.R as the primers. At the same time, vector 2.F and vector 2.R as the primers to obtain pET30a(+)-PT7-RFP vector 1 by PCR. And then construct pET30a(+)-PT7 mutagenesis -RFP with pET30a(+)-PT7-RFP vector 1 and vector 2 by Gibson Assembly. | ||
| Line 27: | Line 16: | ||
<p style="text-align:center;">Fig 2. Site-saturation mutagenesis was conducted to obtain the T7 mutants</p> | <p style="text-align:center;">Fig 2. Site-saturation mutagenesis was conducted to obtain the T7 mutants</p> | ||
| − | <table> | + | <table align="center" class="MsoTableGrid" border="1" cellspacing="0" cellpadding="0" width="567" style="width:425.3pt;margin-left:-7.35pt;border-collapse:collapse;border:none;"> |
| − | vector 1.F CCCGCGAAATTAATACGACTCACTNNNNNGGAATTGTGAGCGGATAAC | + | <tr> |
| − | vector 1.R TACCGCACAGATGCGTAAG | + | <td width="78" valign="top" style="width:58.4pt;border:solid windowtext 1.0pt;padding:0cm 5.4pt 0cm 5.4pt;"><p class="MsoNormal"><span style="font-family:'Times New Roman',serif; font-size:12.0pt; ">vector 1.F<span style="letter-spacing:.15pt; color:black; background:white; "> </span></span></p></td> |
| − | vector 2.F TTCTCCTTACGCATCTGT | + | <td width="489" valign="top" style="width:366.9pt;border:solid windowtext 1.0pt;border-left:none;padding:0cm 5.4pt 0cm 5.4pt;"><p class="MsoNormal"><span style="letter-spacing:.15pt; font-family:'Times New Roman',serif; font-size:12.0pt; color:black; background:white; ">CCCGCGAAATTAATACGACTCACTNNNNNGGAATTGTGAGCGGATAAC</span></p></td> |
| − | vector 2.R AGTGAGTCGTATTAATTTCG</table> | + | </tr> |
| + | <tr> | ||
| + | <td width="78" valign="top" style="width:58.4pt;border:solid windowtext 1.0pt;border-top:none;padding:0cm 5.4pt 0cm 5.4pt;"><p class="MsoNormal"><span style="font-family:'Times New Roman',serif; font-size:12.0pt; ">vector 1.R<span style="color:black; background:white; "> </span></span></p></td> | ||
| + | <td width="489" valign="top" style="width:366.9pt;border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;padding:0cm 5.4pt 0cm 5.4pt;"><p class="MsoNormal"><span style="font-family:'Times New Roman',serif; font-size:12.0pt; color:black; background:white; ">TACCGCACAGATGCGTAAG</span></p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="78" valign="top" style="width:58.4pt;border:solid windowtext 1.0pt;border-top:none;padding:0cm 5.4pt 0cm 5.4pt;"><p class="MsoNormal"><span style="font-family:'Times New Roman',serif; font-size:12.0pt; ">vector 2.F<span style="color:black; background:white; "> </span></span></p></td> | ||
| + | <td width="489" valign="top" style="width:366.9pt;border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;padding:0cm 5.4pt 0cm 5.4pt;"><p class="MsoNormal"><span style="font-family:'Times New Roman',serif; font-size:12.0pt; color:black; background:white; ">TTCTCCTTACGCATCTGT</span></p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="78" valign="top" style="width:58.4pt;border:solid windowtext 1.0pt;border-top:none;padding:0cm 5.4pt 0cm 5.4pt;"><p class="MsoNormal"><span style="font-family:'Times New Roman',serif; font-size:12.0pt; ">vector 2.R<span style="color:black; background:white; "> </span></span></p></td> | ||
| + | <td width="489" valign="top" style="width:366.9pt;border-top:none;border-left:none;border-bottom:solid windowtext 1.0pt;border-right:solid windowtext 1.0pt;padding:0cm 5.4pt 0cm 5.4pt;"><p class="MsoNormal"><span style="font-family:'Times New Roman',serif; font-size:12.0pt; color:black; background:white; ">AGTGAGTCGTATTAATTTCG</span></p></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | |||
<p style="text-align:center;">Table 1: Primers for site-saturation mutagenesis</p> | <p style="text-align:center;">Table 1: Primers for site-saturation mutagenesis</p> | ||
| Line 37: | Line 40: | ||
T7 mutants were preliminarily selected by fluorescence of colonies grown on LB agar plates with Lactose. These mutants were inoculated in 2ml LB medium and cultured at 37 °C with agitation at 250 rpm for 12 h. Then, 1:100 inoculate to 200μl LB medium (with 0.2mM IPTG) in 96 - well plates at 30°C with agitation at 250 rpm for 16 h. The excitation wavelength was set to 553 nm, and the emission wavelength was set to 583 nm. The optical density of bacteria at 600 nm was detected with a spectrophotometer. | T7 mutants were preliminarily selected by fluorescence of colonies grown on LB agar plates with Lactose. These mutants were inoculated in 2ml LB medium and cultured at 37 °C with agitation at 250 rpm for 12 h. Then, 1:100 inoculate to 200μl LB medium (with 0.2mM IPTG) in 96 - well plates at 30°C with agitation at 250 rpm for 16 h. The excitation wavelength was set to 553 nm, and the emission wavelength was set to 583 nm. The optical density of bacteria at 600 nm was detected with a spectrophotometer. | ||
Finally, we have screened 8 different strength T7 mutants. The following figure is fluorescence/OD600 of them. | Finally, we have screened 8 different strength T7 mutants. The following figure is fluorescence/OD600 of them. | ||
| − | + | <center>[[File:T--SCUT China--Pt7 site fig.jpeg|500px]]<br> | |
| − | <p style="text-align:center;">Fig. 2: fluorescence/OD600 of T7 mutants</p> | + | <p style="text-align:center;">Fig. 2: fluorescence/OD600 of T7 mutants</p></center> |
| − | <strong>Reference: </strong> | + | <h2><strong>Reference: </strong></h2> |
[1] Nie, Z., Luo, H., Li, J. et al. Appl Biochem Biotechnol (2019). https://doi.org/10.1007/s12010-019-03113-y | [1] Nie, Z., Luo, H., Li, J. et al. Appl Biochem Biotechnol (2019). https://doi.org/10.1007/s12010-019-03113-y | ||
Latest revision as of 03:55, 22 October 2019
T7 promoter variants family member
Usage and Biology:
PT7 is the most well-known inducible promoter with high transcriptional strength[1]. T7 RNAP is a single-subunit RNA polymerase that is a strong driver of transcription. It is functionally orthogonal to most hosts, acting only on its cognate promoter, PT7. In order to achieve a larger scale and more accurate regulation range, we have improved T7 promoters (BBa_j64997)with different strength providing more options for the precise regulation.
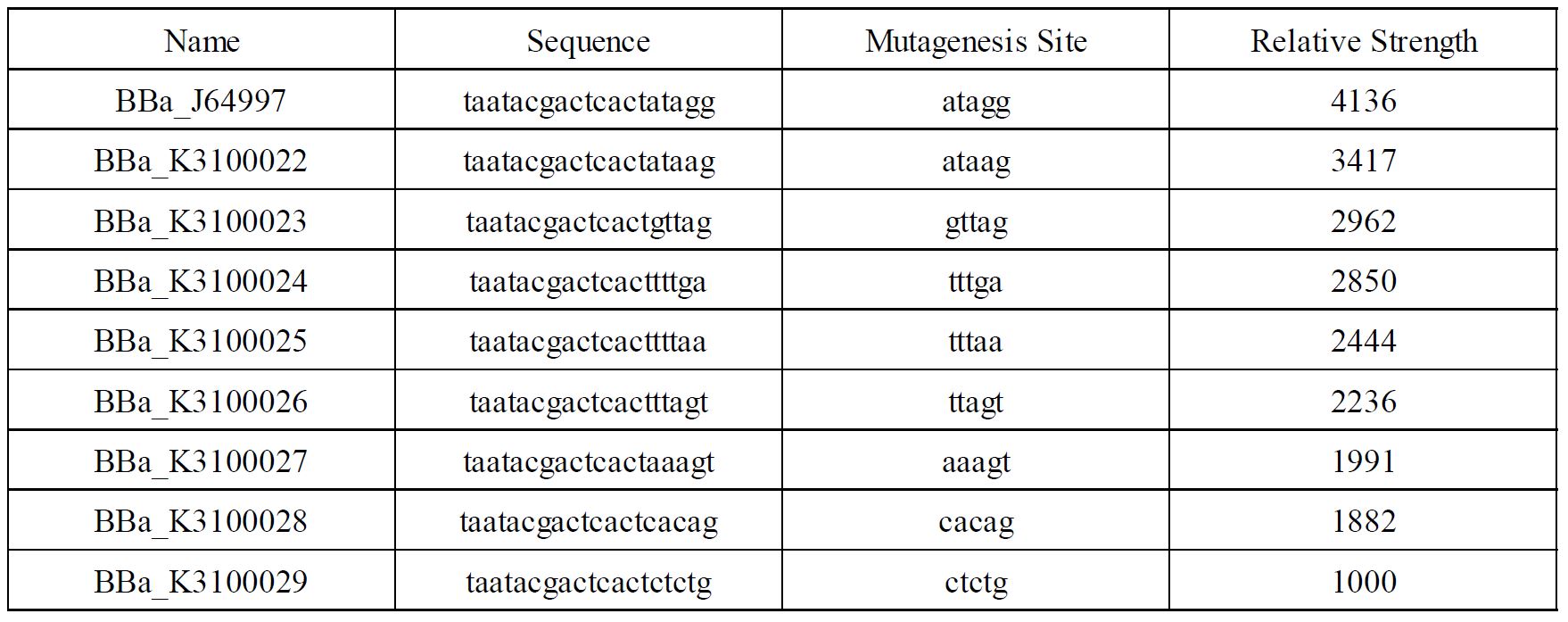
Fig1. T7 promoters with different strength
Characterization:
1. Construction of the T7 Promoter Library
A T7 promoter library was constructed through site-saturation mutagenesis, and an efficient screening method based on GFP fluorescence detection was established.
The site-saturation mutagenesis DNA fragment pET30a(+)-PT7-RFP vector 1 was obtained using the plasmid pET30a(+)-PT7-RFP as the template and vector 1.F (the sequence containing the mutation in promoter)and vector 1.R as the primers. At the same time, vector 2.F and vector 2.R as the primers to obtain pET30a(+)-PT7-RFP vector 1 by PCR. And then construct pET30a(+)-PT7 mutagenesis -RFP with pET30a(+)-PT7-RFP vector 1 and vector 2 by Gibson Assembly.
Fig 2. Site-saturation mutagenesis was conducted to obtain the T7 mutants
vector 1.F |
CCCGCGAAATTAATACGACTCACTNNNNNGGAATTGTGAGCGGATAAC |
vector 1.R |
TACCGCACAGATGCGTAAG |
vector 2.F |
TTCTCCTTACGCATCTGT |
vector 2.R |
AGTGAGTCGTATTAATTTCG |
Table 1: Primers for site-saturation mutagenesis
2. T7 Mutant Screening and Determination of GFP Fluorescence
T7 mutants were preliminarily selected by fluorescence of colonies grown on LB agar plates with Lactose. These mutants were inoculated in 2ml LB medium and cultured at 37 °C with agitation at 250 rpm for 12 h. Then, 1:100 inoculate to 200μl LB medium (with 0.2mM IPTG) in 96 - well plates at 30°C with agitation at 250 rpm for 16 h. The excitation wavelength was set to 553 nm, and the emission wavelength was set to 583 nm. The optical density of bacteria at 600 nm was detected with a spectrophotometer.
Finally, we have screened 8 different strength T7 mutants. The following figure is fluorescence/OD600 of them.
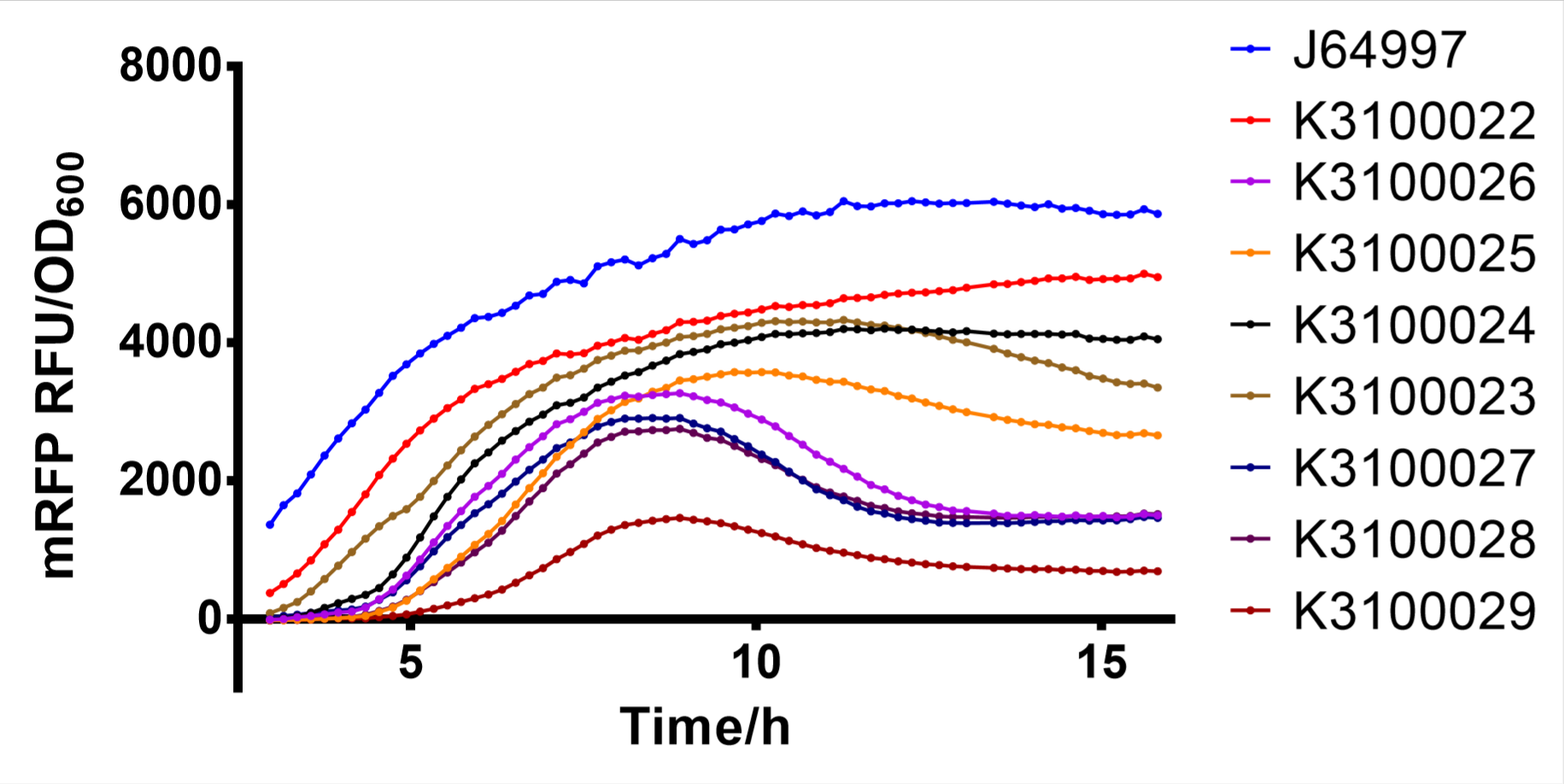
Fig. 2: fluorescence/OD600 of T7 mutants
Reference:
[1] Nie, Z., Luo, H., Li, J. et al. Appl Biochem Biotechnol (2019). https://doi.org/10.1007/s12010-019-03113-y
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]

