Difference between revisions of "Part:BBa K2906000"
| (21 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
<partinfo>BBa_K2906000 short</partinfo> | <partinfo>BBa_K2906000 short</partinfo> | ||
| − | '''This is an improved part of <partinfo>BBa_I746909</partinfo>. By adding a Tet promoter, we are ensuring that sfGFP is tightly controlled, and expressing a higher level of fluorescence in the initial hours after induction [https:// | + | '''This is an improved part of <partinfo>BBa_I746909</partinfo>. By adding a Tet promoter, we are ensuring that sfGFP is tightly controlled, and expressing a higher level of fluorescence in the initial hours after induction. For full quantitative comparison visit [https://2019.igem.org/Team:Manchester/Improve Team Manchester Wiki]''' |
| − | Superfolder GFP also known as sfGFP is a GFP-derived green fluorescent protein. (Pédelacq et al., 2006). GFP is a protein isolated from the jellyfish Aequorea Victoria that exhibits green fluorescence when exposed to light in the blue to the ultraviolet range (Prendergast and Mann, 1978; Tsien, 1998). A series of mutations performed by Pédelacq et al. obtained a GFP variant able to rapidly fold and mature. This ultimately | + | Superfolder GFP also known as sfGFP is a GFP-derived green fluorescent protein. (Pédelacq et al., 2006). GFP is a protein isolated from the jellyfish Aequorea Victoria that exhibits green fluorescence when exposed to light in the blue to the ultraviolet range (Prendergast and Mann, 1978; Tsien, 1998). A series of mutations performed by Pédelacq et al. obtained a GFP variant able to rapidly fold and mature. This ultimately leads to enhanced fluorescence intensity (Frenzel et al., 2018). This composite part is made of sfGFP (<partinfo>BBa_K1321337</partinfo>) expressed by a Tet promoter (<partinfo>BBa_R0040</partinfo>). The protein is able to express colour and auto secrets to the media. |
__FORCETOC__ | __FORCETOC__ | ||
| Line 16: | Line 16: | ||
[[File:SfGFP RFU OD.png|750px|center]] | [[File:SfGFP RFU OD.png|750px|center]] | ||
| − | '''Figure 1.''' The plot shows the mean RFU/OD from three replicates of each construct expressed in ''E. coli'' DH5⍺ and BL21(DE3). The OD was measured at 600 nm and GFP fluorescence was measured at Ex ƛ 485, Em ƛ 510, every 15 minutes for 13 hours. The RFU values were normalised by the OD and the triplicates averaged. All values have been blank-corrected. A total of 52 recordings were made per well, with three | + | '''Figure 1.''' The plot shows the mean RFU/OD from three replicates of each construct expressed in ''E. coli'' DH5⍺ and BL21(DE3). The OD was measured at 600 nm and GFP fluorescence was measured at Ex ƛ 485, Em ƛ 510, every 15 minutes for 13 hours. The RFU values were normalised by the OD and the triplicates averaged. All values have been blank-corrected. A total of 52 recordings were made per well, with three technical replicates per construct. 'Colour Alone' represents this part, pTet + sfGFP. The values suggest that sfGFP is expressed at very high levels in both DH5a and BL21(DE3). |
| + | |||
| + | <div style='text-align: left;'>'''SDS:</div> | ||
| + | |||
| + | We also wanted to visualise if the protein was being overexpressed and secreted across the bacterial membrane. We first calculated the expected size of sfGFP of our desired construct: 26.8 kDa molecular weight. Then samples were run on 10% gels following SDS-PAGE protocol. Below we show the gel containing samples from the supernatant, which proves that this construct indeed auto secrets. | ||
| + | |||
| + | [[File:T--Manchester--Parts--SDS-Supernatant-sfGFP.png|350px|center]] | ||
| − | |||
| − | |||
===Qualitative Data:=== | ===Qualitative Data:=== | ||
| + | |||
| + | '''Fluorescence Microscopy:''' | ||
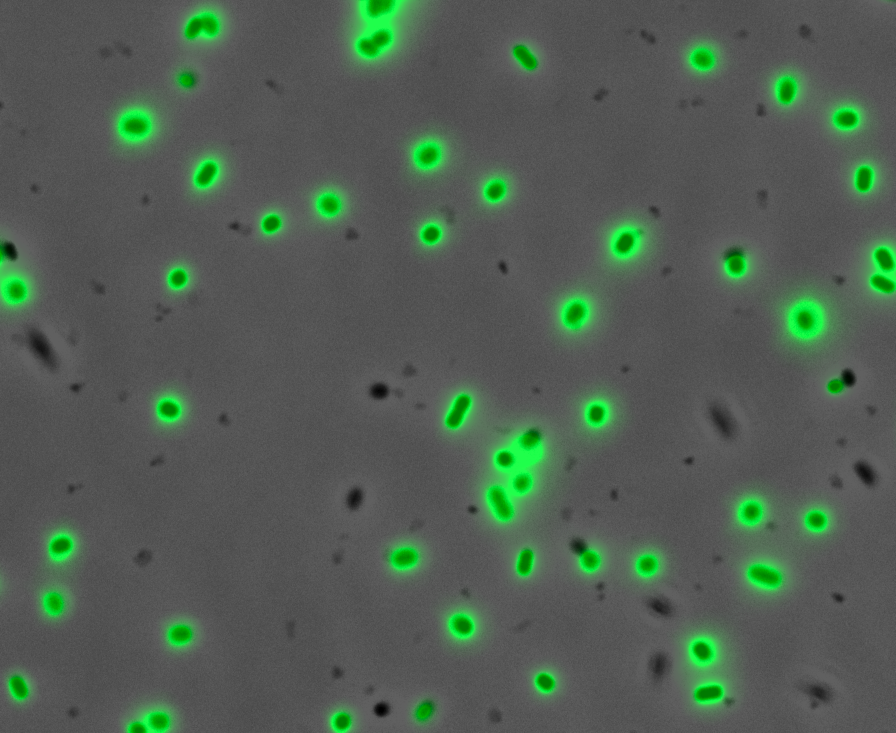
| + | After quantifying the data for sfGFP we decided to perform fluorescence microscopy. This would allow us to see the percentage of bacteria in a sample producing colour, as well as some phenotypic characteristics of the bacteria with the construct. Below you can see two images, the negative control, which was ''E. coli'' TOP10, showing very limited background GFP fluorescence, and BBa_K2906000, clearly showing the expression and functionality of sfGFP with Tet promoter. | ||
| + | |||
| + | <br /><div style='text-align: left;'>'''Negative Control:'''</div> <div style='text-align: right;'>'''BBa_K2906000:'''</div> | ||
| + | [[File:T--Manchester--Colours--FM--NegO.png|440px|left]] [[File:FM_Gold_Colour_Alone_Overlay.png|440px|right]]<br /><br /><br /> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br /><br />'''Figure 2. Fluorescent microscopy overlaid images of phase contrast and GFP filter.''' a) Negative control (''E. coli'' TOP10) showing very slight background readings for fluorescence. b) sfGFP with Tet (BBa_K2906000) showing significant levels of fluorescence in most bacteria. The images above are composite overlaid images made from phase contrast and GFP-filter captures. They were processed using ImageJ. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | '''Colour:''' | ||
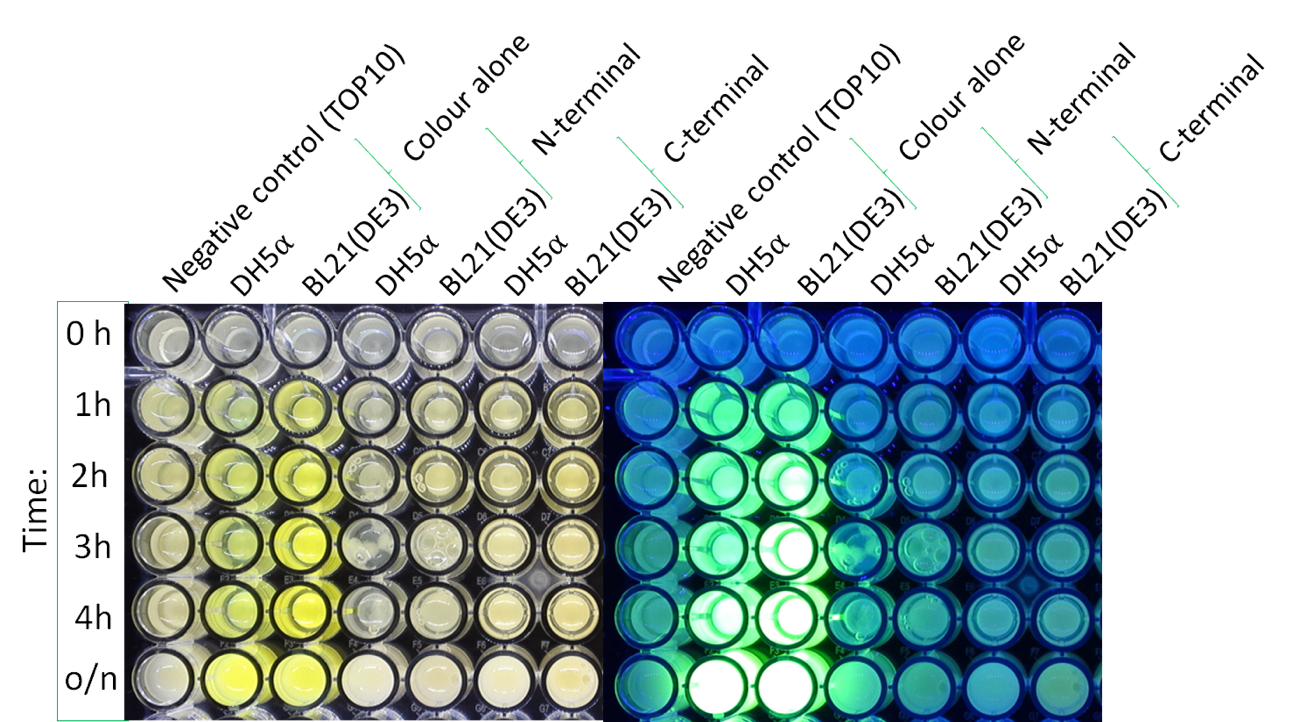
| + | The image below shows the visible colour of the pellet obtained under normal light and UV light. This was done at five different time points after induction with 100 nM of anhydrotetracycline. sfGFP with Tet is labelled <nowiki>''Colour Alone''</nowiki> and can be seen present in both DH5a and BL21. | ||
[[File:SfGFP Colour Pellets.png|750px|center]] | [[File:SfGFP Colour Pellets.png|750px|center]] | ||
| − | This is an essential new part, as sfGFP is a very common reporter gene. Additionally, we show in our wiki [https://2019.igem.org/Team:Manchester] how this is an improved part over the sfGFP provided by iGEM with a T7 promoter. We hope that this new part can be useful for future teams! | + | This is an essential new part, as sfGFP is a very common reporter gene. Additionally, we show in our wiki [https://2019.igem.org/Team:Manchester/Improve Team Manchester] how this is an improved part over the sfGFP provided by iGEM with a T7 promoter. We hope that this new part can be useful for future teams! |
===References:=== | ===References:=== | ||
Latest revision as of 16:35, 21 October 2019
sfGFP with Tet Promoter
This is an improved part of BBa_I746909. By adding a Tet promoter, we are ensuring that sfGFP is tightly controlled, and expressing a higher level of fluorescence in the initial hours after induction. For full quantitative comparison visit Team Manchester Wiki
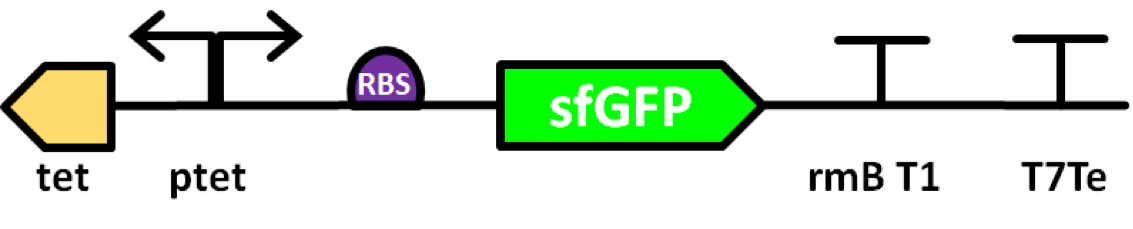
Superfolder GFP also known as sfGFP is a GFP-derived green fluorescent protein. (Pédelacq et al., 2006). GFP is a protein isolated from the jellyfish Aequorea Victoria that exhibits green fluorescence when exposed to light in the blue to the ultraviolet range (Prendergast and Mann, 1978; Tsien, 1998). A series of mutations performed by Pédelacq et al. obtained a GFP variant able to rapidly fold and mature. This ultimately leads to enhanced fluorescence intensity (Frenzel et al., 2018). This composite part is made of sfGFP (BBa_K1321337) expressed by a Tet promoter (BBa_R0040). The protein is able to express colour and auto secrets to the media.
Characterisation:
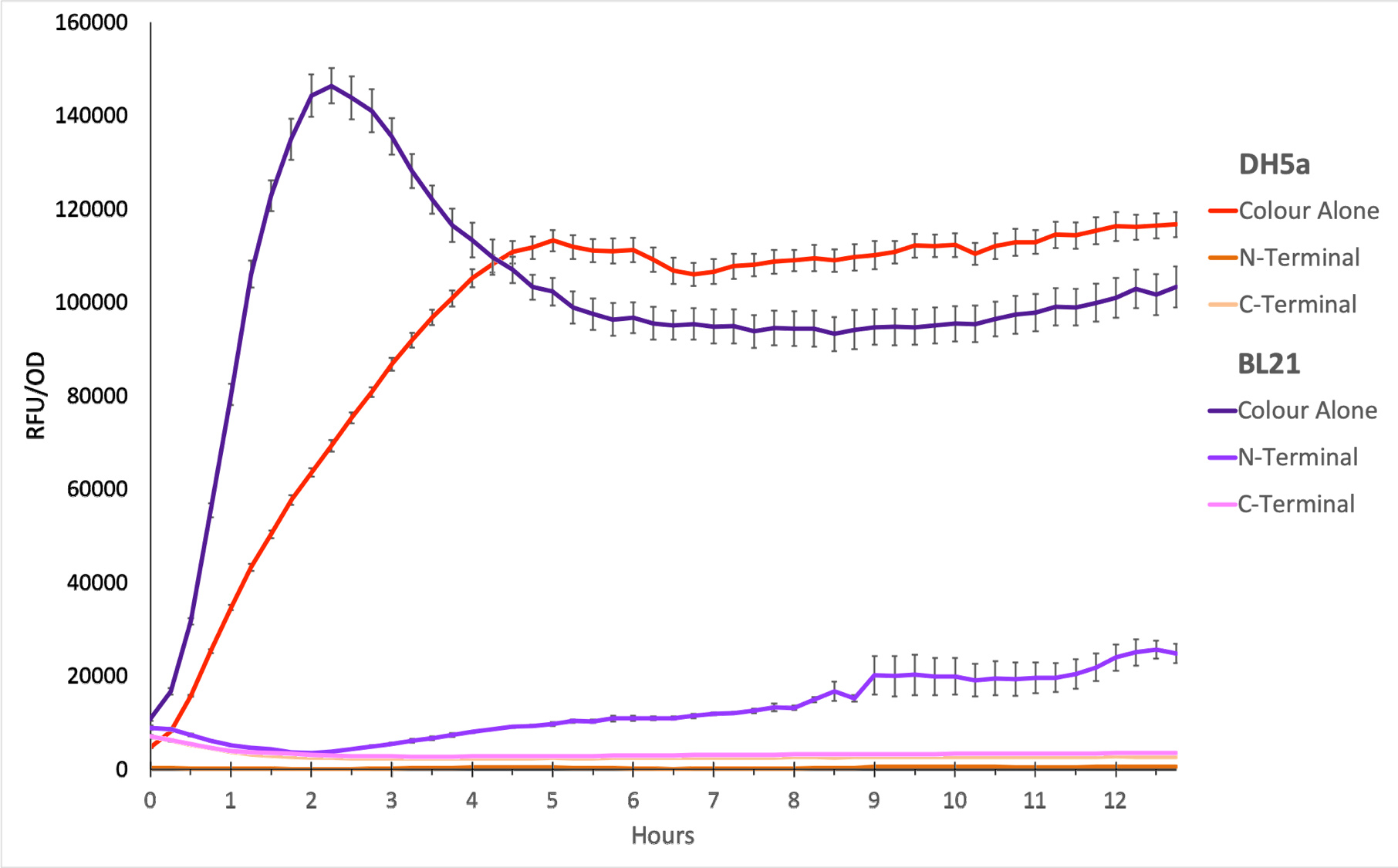
To show whether this part worked, we decided to measure fluorescence, normalised by OD. Optical Density values were measured at 600 nm. We grew the cells to an OD600 of ~0.6 and induced them with 100 nM of anhydrotetracycline. Then, they were loaded in triplicates into a 96-well plate and left overnight. For OD values, the blank was LB media, and for RFU the blank was E. coli TOP10 since it does not express any colour. The values were individually normalised by dividing RFU/OD and then averaged to plot the mean against time. An RFU value of 0 corresponds to baseline E. coli TOP10 measurements.
Figure 1. The plot shows the mean RFU/OD from three replicates of each construct expressed in E. coli DH5⍺ and BL21(DE3). The OD was measured at 600 nm and GFP fluorescence was measured at Ex ƛ 485, Em ƛ 510, every 15 minutes for 13 hours. The RFU values were normalised by the OD and the triplicates averaged. All values have been blank-corrected. A total of 52 recordings were made per well, with three technical replicates per construct. 'Colour Alone' represents this part, pTet + sfGFP. The values suggest that sfGFP is expressed at very high levels in both DH5a and BL21(DE3).
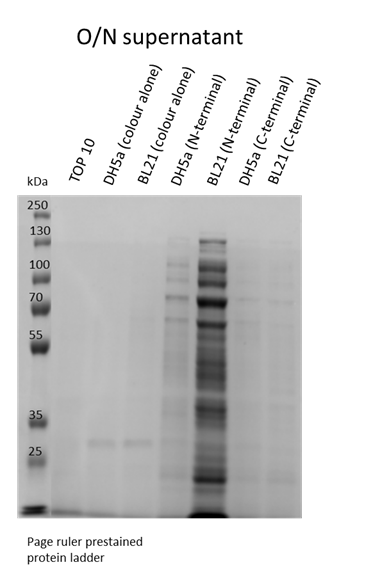
We also wanted to visualise if the protein was being overexpressed and secreted across the bacterial membrane. We first calculated the expected size of sfGFP of our desired construct: 26.8 kDa molecular weight. Then samples were run on 10% gels following SDS-PAGE protocol. Below we show the gel containing samples from the supernatant, which proves that this construct indeed auto secrets.
Qualitative Data:
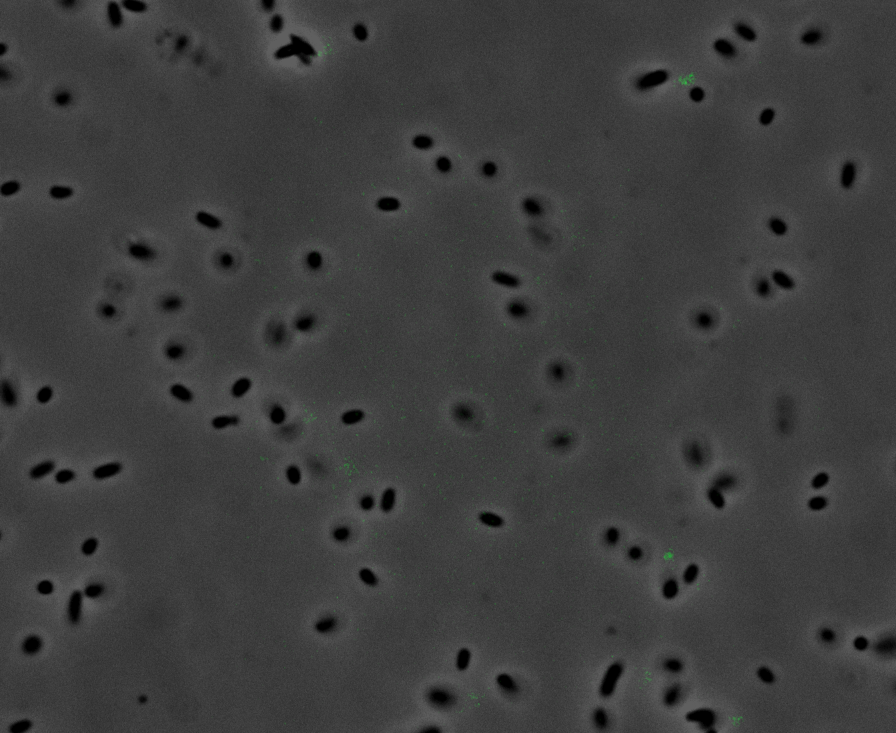
Fluorescence Microscopy: After quantifying the data for sfGFP we decided to perform fluorescence microscopy. This would allow us to see the percentage of bacteria in a sample producing colour, as well as some phenotypic characteristics of the bacteria with the construct. Below you can see two images, the negative control, which was E. coli TOP10, showing very limited background GFP fluorescence, and BBa_K2906000, clearly showing the expression and functionality of sfGFP with Tet promoter.
Figure 2. Fluorescent microscopy overlaid images of phase contrast and GFP filter. a) Negative control (E. coli TOP10) showing very slight background readings for fluorescence. b) sfGFP with Tet (BBa_K2906000) showing significant levels of fluorescence in most bacteria. The images above are composite overlaid images made from phase contrast and GFP-filter captures. They were processed using ImageJ.
Colour:
The image below shows the visible colour of the pellet obtained under normal light and UV light. This was done at five different time points after induction with 100 nM of anhydrotetracycline. sfGFP with Tet is labelled ''Colour Alone'' and can be seen present in both DH5a and BL21.
This is an essential new part, as sfGFP is a very common reporter gene. Additionally, we show in our wiki Team Manchester how this is an improved part over the sfGFP provided by iGEM with a T7 promoter. We hope that this new part can be useful for future teams!
References:
Frenzel, E., Legebeke, J., Van Stralen, A., Van Kranenburg, R. and Kuipers, O. P. (2018) ‘In vivo selection of sfGFP variants with improved and reliable functionality in industrially important thermophilic bacteria’, Biotechnology for Biofuels. BioMed Central Ltd., 11(1). doi: 10.1186/s13068-017-1008-5.
Pédelacq, J. D., Cabantous, S., Tran, T., Terwilliger, T. C. and Waldo, G. S. (2006) ‘Engineering and characterization of a superfolder green fluorescent protein’, Nature Biotechnology, 24(1), pp. 79–88. doi: 10.1038/nbt1172.
Prendergast, F. G. and Mann, K. G. (1978) ‘Chemical and Physical Properties of Aequorin and the Green Fluorescent Protein Isolated from Aequorea forskålea†’, Biochemistry, 17(17), pp. 3448–3453. doi: 10.1021/bi00610a004.
Tsien, R. Y. (1998) ‘THE GREEN FLUORESCENT PROTEIN’, Annual Review of Biochemistry. Annual Reviews, 67(1), pp. 509–544. doi: 10.1146/annurev.biochem.67.1.509.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 92