Difference between revisions of "Part:BBa K2997000"
(→Expression in E. coli) |
(→TAL and TyrP Functional Assay) |
||
| (2 intermediate revisions by the same user not shown) | |||
| Line 16: | Line 16: | ||
<br> | <br> | ||
</html> | </html> | ||
| − | Fig. 1. Confirmation of BBa_K2997000 by double digestion. M: Marker; Lane 1: BBa_K2997000, arrow indicates | + | Fig. 1. Confirmation of BBa_K2997000 by double digestion. M: Marker; Lane 1: BBa_K2997000, arrow indicates <i>P<sub>tyrP</sub></i>-<i>tyrP</i> insert at 1307bp; Lane 2: pSB4A3-J04450 (negative control). |
===TAL and TyrP Functional Assay=== | ===TAL and TyrP Functional Assay=== | ||
| − | To confirm the protein activity of TAL and TyrP, we performed a functional test using n-octanol extraction method (https://2019.igem.org/Team:NCKU_Tainan/Protocols), which was previously proposed by iGEM Uppsala 2013 and has been verified by HPLC[ | + | To confirm the protein activity of TAL and TyrP, we performed a functional test using <i>n</i>-octanol extraction method (https://2019.igem.org/Team:NCKU_Tainan/Protocols), which was previously proposed by iGEM Uppsala 2013 and has been verified by HPLC<sup>[1]</sup>. The <i>p</i>-Coumaric acid concentration was measured through the absorbance value at 310nm wavelength under Nanodrop UV-Vis wavelength. The standard curve of <i>p</i>-Coumaric acid was drawn in Fig.2 to determine the relationship between <i>p</i>-Coumaric acid concentrations and its 310nm arbitrary unit (a.u). Our samples with TAL constructs were then mapped onto the standard curve, to know how much <i>p</i>-Coumaric acid is being produced. |
<html> | <html> | ||
<br> | <br> | ||
| Line 29: | Line 29: | ||
Fig. 2. The standard curve of <i>p</i>-Coumaric acid concentration in correlation with absorbance at 310nm, which is provided by genetic <i>E. coli</i> Nissle in LB broth after 48 hours. | Fig. 2. The standard curve of <i>p</i>-Coumaric acid concentration in correlation with absorbance at 310nm, which is provided by genetic <i>E. coli</i> Nissle in LB broth after 48 hours. | ||
| − | Using this assay, We can further improve the conversion of tyrosine into <i>p</i>-Coumaric acid by adding a tyrosine transporter (BBa_K2997000). As seen in Fig | + | Using this assay, We can further improve the conversion of tyrosine into <i>p</i>-Coumaric acid by adding a tyrosine transporter (BBa_K2997000). As seen in Fig. 2, when tyrosine transporter is added, the production of <i>p</i>-Coumaric acid is significantly higher than when it is not added. When tyrosine transporter (BBa_K2997000) is introduced into <i>E. coli</i> Nissle with TAL constructs containing native RBS (BBa_K2997009) and B0034 RBS (BBa_K2997010), conversion of tyrosine into <i>p</i>-Coumaric acid is increased by 1.44-fold and 1.31-fold respectively. |
<html> | <html> | ||
Latest revision as of 12:32, 20 October 2019
Tyrosine transporter (tyrP)
Background
It’s a gene that encodes for tyrosine transporter, which is involved in transporting tyrosine across the cytoplasmic membrane. Hence, we used PCR to amplify this gene from E. coli MG1655 and incorporated it to our E. coli, allowing it to take in more tyrosine.
Expression in E. coli
We amplified the tyrP gene and its native promoter from E. coli MG1655 and clone it into pSB4A3. Then transformed the plasmid into DH5α and E. coli Nissle 1917. Plasmid extraction after the colony formed and follow up by enzyme digestion to confirm the insertion was successful.

Fig. 1. Confirmation of BBa_K2997000 by double digestion. M: Marker; Lane 1: BBa_K2997000, arrow indicates PtyrP-tyrP insert at 1307bp; Lane 2: pSB4A3-J04450 (negative control).
TAL and TyrP Functional Assay
To confirm the protein activity of TAL and TyrP, we performed a functional test using n-octanol extraction method (https://2019.igem.org/Team:NCKU_Tainan/Protocols), which was previously proposed by iGEM Uppsala 2013 and has been verified by HPLC[1]. The p-Coumaric acid concentration was measured through the absorbance value at 310nm wavelength under Nanodrop UV-Vis wavelength. The standard curve of p-Coumaric acid was drawn in Fig.2 to determine the relationship between p-Coumaric acid concentrations and its 310nm arbitrary unit (a.u). Our samples with TAL constructs were then mapped onto the standard curve, to know how much p-Coumaric acid is being produced.
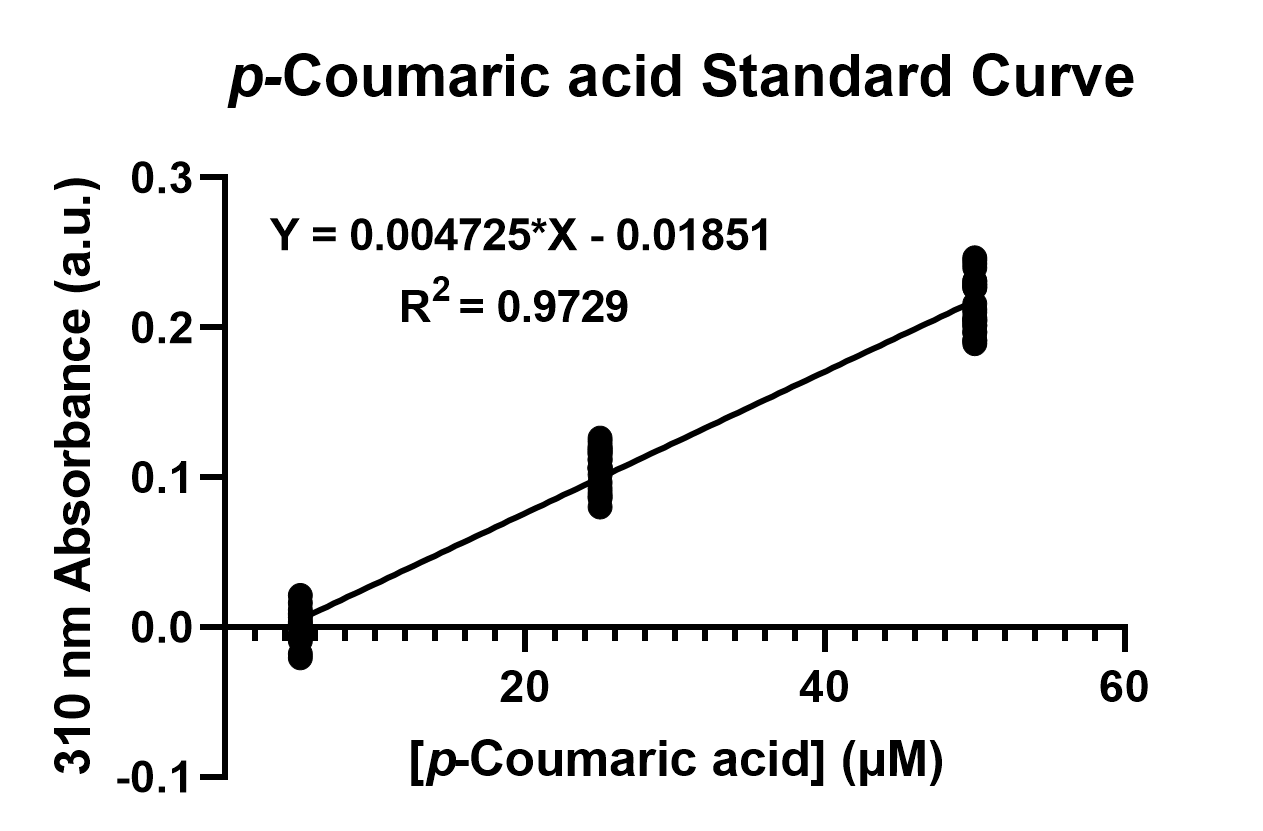
Fig. 2. The standard curve of p-Coumaric acid concentration in correlation with absorbance at 310nm, which is provided by genetic E. coli Nissle in LB broth after 48 hours.
Using this assay, We can further improve the conversion of tyrosine into p-Coumaric acid by adding a tyrosine transporter (BBa_K2997000). As seen in Fig. 2, when tyrosine transporter is added, the production of p-Coumaric acid is significantly higher than when it is not added. When tyrosine transporter (BBa_K2997000) is introduced into E. coli Nissle with TAL constructs containing native RBS (BBa_K2997009) and B0034 RBS (BBa_K2997010), conversion of tyrosine into p-Coumaric acid is increased by 1.44-fold and 1.31-fold respectively.
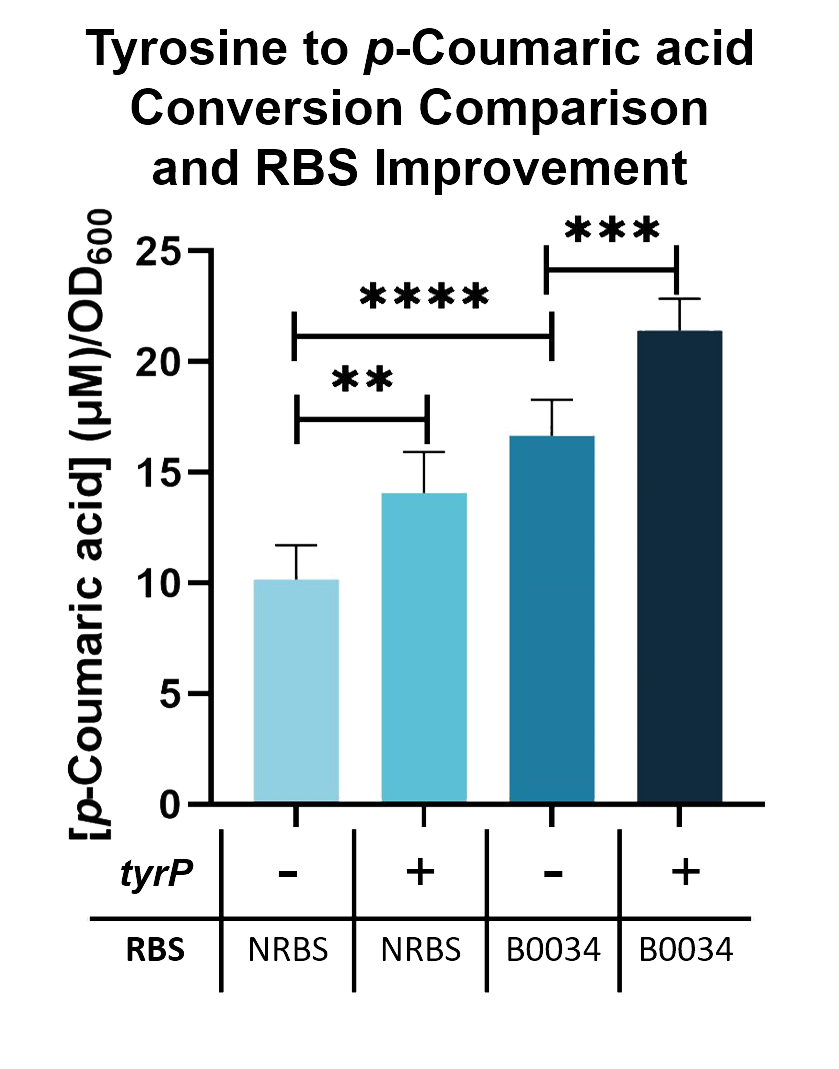
Fig. 3. p-Coumaric acid/O.D.600 levels of E. coli Nissle with TAL and tyrP in LB with 1mM tyrosine
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 278
- 1000COMPATIBLE WITH RFC[1000]
Reference
[1] Wookey, P. J., & Pittard, A. J. (1988). DNA sequence of the gene (tyrP) encoding the tyrosine-specific transport system of Escherichia<i/> coli. Journal of Bacteriology, 170(10), 4946–4949. doi: 10.1128/jb.170.10.4946-4949.1988
