Difference between revisions of "Part:BBa K2959005"
(→Characterization of Expressible Pinus sylvestris Defensin 1 with Erv1p) |
(→Antifungal Assay) |
||
| (38 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K2959005 short</partinfo> | <partinfo>BBa_K2959005 short</partinfo> | ||
<p align="justify"> | <p align="justify"> | ||
This composite part consists of a lacI regulated promoter, ribosome binding site, a coding sequence for <i>Pinus sylvestris</i> Defensin 1 as a fusion protein with a 6x His-Tag, a second ribosome binding site, a coding sequence for Erv1p, and a double terminator. This construct allows the expression of PsDef1, an antifungal peptide from Scots pine seeds, in <i>E. coli</i>. Expression can be positively regulated by the addition of IPTG or lactose thanks to the lacl regulated promoter. The fusion of PsDef1 to a 6x His-Tag is intended for the purification of the peptide by immobilized metal affinity chromatography. | This composite part consists of a lacI regulated promoter, ribosome binding site, a coding sequence for <i>Pinus sylvestris</i> Defensin 1 as a fusion protein with a 6x His-Tag, a second ribosome binding site, a coding sequence for Erv1p, and a double terminator. This construct allows the expression of PsDef1, an antifungal peptide from Scots pine seeds, in <i>E. coli</i>. Expression can be positively regulated by the addition of IPTG or lactose thanks to the lacl regulated promoter. The fusion of PsDef1 to a 6x His-Tag is intended for the purification of the peptide by immobilized metal affinity chromatography. | ||
| − | + | <br> | |
Since PsDef1 is a peptide with four disulfide bonds<sup>1</sup>, a modification hard to replicate on prokaryotic expression systems, the construct allows for the co-expression of the peptide with a truncated version of Erv1p. This is a protein from Saccharomyces cerevisiae capable of catalyzing the formation of disulfide bonds<sup>2</sup>. | Since PsDef1 is a peptide with four disulfide bonds<sup>1</sup>, a modification hard to replicate on prokaryotic expression systems, the construct allows for the co-expression of the peptide with a truncated version of Erv1p. This is a protein from Saccharomyces cerevisiae capable of catalyzing the formation of disulfide bonds<sup>2</sup>. | ||
| − | + | </p> | |
| + | <br> | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| − | PsDef1 is present in <i>Pinus sylvestris</i> seeds, vegetative and generative organs. The defensin encodes a protein of 83 amino acids in length, whose first 33 amino acids correspond to the ‘N-terminal signal peptide, it also has eight highly conserved cysteines residues that are predicted to form four disulfide bonds C3–C49, C14–C34, C20–C43, and C24–C45<sup>1</sup>, which stabilize its structure making it resistant to environmental changes<sup>2</sup>. Its conformation consists in a cysteine-stabilized αβ-motif (CSαβ) with a prominent α-helix and a triple-stranded antiparallel β-sheet that is stabilized by the 4 disulfide bonds<sup>1</sup>. | + | <p align="justify"> |
| + | PsDef1 is present in <i>Pinus sylvestris</i> seeds, vegetative and generative organs. The defensin encodes a protein of 83 amino acids in length, whose first 33 amino acids correspond to the ‘N-terminal signal peptide, it also has eight highly conserved cysteines residues that are predicted to form four disulfide bonds C3–C49, C14–C34, C20–C43, and C24–C45<sup>1</sup>, which stabilize its structure making it resistant to environmental changes<sup>2</sup>. Its conformation consists in a cysteine-stabilized αβ-motif (CSαβ) with a prominent α-helix and a triple-stranded antiparallel β-sheet that is stabilized by the 4 disulfide bonds<sup>1</sup>.</p> | ||
| + | <p align="justify"> | ||
| + | Endogenous PsDef1 causes morphological changes in fungi mycelium when interacting with the sphingolipid membrane on fungal cell, disrupting the membrane integrity. Antifungal activity towards phytopathogenic fungi <i>Botrytis cinerea</i>, <i>Fusarium oxysporum</i>, <i>Fusarium solani</i>, and <i>Heterobasidion annosum</i> has been demonstrated; as well as its antimicrobial activity against Gram-positive (<i>Bacillus pumilus</i>) and Gram-negative bacteria (<i>Pectobacterium carotovorum</i>, <i>Pseudomonas fluorescens</i>)<sup>1</sup>.</p> | ||
| + | |||
| + | <p align="justify"> | ||
| + | For a successful expression in <i>E. coli</i>, it is indispensable to avoid interactions that result in the aggregation of folding intermediates. Disulfide bonds can be involved in the structural, catalytic and signaling roles of the protein, but the formation of disulfide bonds can have certain problems that can cause misfolding, aggregation and low yields during the production of the protein<sup>4</sup>. The usage of Erv1p has been proposed as a mechanism for the formation of disulfide bonds in recombinant proteins in prokaryotic expression systems. It has been shown that, thanks to its ability to form disulfide bonds <i>de novo</i>, co-expression of Erv1p allows the proper formation of disulfide bonded proteins in the cytoplasm of <i>E. coli</i><sup>5</sup>.</p> | ||
| + | |||
| + | <p align="justify"> | ||
| + | The ERV1 gene from <i>Saccharomyces cerevisiae</i> encodes for a 189 amino acids protein, Erv1p, that is involved in different processes of mitochondrial biogenesis and maintenance<sup>2, 6</sup>. The protein shows a flavin-linked sulfhydryl oxidase enzymatic activity linked to the carboxy-terminal domain. Thus, Erv1p is capable of oxidizing thiol groups in proteins and catalyzing disulfide bond formation. A 15 kDa truncated version of Erv1p, consisting of the 117 amino acid residues carboxy-terminal domain, shows a similar or improved sulfhydryl oxidase activity compared to the full length protein<sup>2</sup>.</p> | ||
| + | <br> | ||
| + | ==<b>Characterization of Expressible Pinus sylvestris Defensin 1 with Erv1p</b>== | ||
| + | <p align= "justify"> | ||
| + | Our DNA sequence PsDef1-Erv1p was synthesized by IDT®️ with the Biobrick prefix and suffix flanking the composite part. This made possible the correct digestion with restriction enzymes EcoRI-HF and PstI. After the digestion, ligation was performed with T7 ligase in order to place our construct into the pSB1C3 linearized backbone with chloramphenicol resistance, which was previously digested with the same restriction enzymes. Using the SnapGene®️ software, we could model our ligated expression plasmid, and the final part resulted in a sequence of 3,106 bp. Thereupon, <i>Escherichia coli</i> SHuffle was transformed by heat shock for following antibiotic selection of clones. | ||
</p> | </p> | ||
| + | <center>[[File:T--Tec-Chihuahua--PsDef-Erv1pff.png|400px]]</center> | ||
| + | <center><b>Figure 1.</b>SnapGene®️ map of BioBrick BBa_K2959005</center> | ||
| + | |||
| + | <p align="justify"> | ||
| + | The next step was to amplify our BioBrick sequence through colony PCR performed upon our transformed cells to confirm the presence of our expression plasmid inside of our chassis. With the help of the specific forward Biobrick prefix [BBa_G1004] and the specific reverse Biobrick suffix [BBa_G1005], we were able to amplify our sequence exclusively. Through an agarose gel we confirmed the correct transformation. The PCR action from SnapGene®️ was used to predict the size of the amplified sequences which resulted in a size of 1,095 for PsDef1+Erv1p.</p> | ||
| + | <center>[[File:T--Tec-Chihuahua--psdef1.pcr..png|600px]] | ||
| + | [[File:T--Tec-Chihuahua--ELECTROFORESISPsDefE.jpeg|200px]]</center> | ||
| + | <center><b>Figure 2.</b>(On the left) SnapGene®️ amplification through PCR of BBa_K2959005. (On the right) Agarose gel electrophoresis of BBa_K2959005 compared with NEB Quick-Load Purple 1Kb Plus DNA Ladder, where the highlighted band corresponds to approximately 1,095 bp.</center> | ||
| + | <br> | ||
| + | ===Protein production=== | ||
| + | <b>IPTG Induction and Extraction</b> | ||
<br> | <br> | ||
<p align="justify"> | <p align="justify"> | ||
| − | + | Following the construction of the BioBrick, it was necessary to induce protein production. Production of PsDef1 was possible under Lacl promoter when induced with 0.2 or 0.4 mM IPTG at 30°C and 225 rpm. This was followed by protein extraction by lysis solution to which lysozyme was added in order to obtain our soluble peptides. | |
| + | </p> | ||
| + | ===Antifungal Assay=== | ||
| + | <p align="justify"> | ||
| + | <br> | ||
| + | In order to test the antifungal activity of our peptides as well as prove their viability as a mechanism to inhibit <i>Verticillium dahliae</i>, an antifungal susceptibility test on a 96 well plate was carried out by measuring absorbance at 405 nm, wavelength used in standardized protocols to measure growth of filamentous fungi<sup>7</sup>. Due to a lack of time, we couldn’t reach the experimental stage of the project of peptide purification, so experiments were made using soluble protein extracts from our transformed cells’ lysates. Different dilutions of the extracts were prepared which were applied to a spore suspension of <i>V. dahliae</i>. Dilutions of the extracts of untransformed cells were used as controls to prove that inhibition was the result of the peptides and not any other protein contained within the extract. Soluble proteins from <i>E. coli</i> SHuffle were used as control for the PsDef1 extract. Concentrations of extracts with peptides were equalized to their controls. Table 1 details the concentrations of each dilution. | ||
</p> | </p> | ||
| + | <center> <b>Table 1. </b>Protein concentration of soluble protein extract of transformed cells induced to produce PsDef1 and untransformed SHuffle control. </center> | ||
| + | <table class="lab-table" style="margin-left: auto; margin-right: auto;"> | ||
| + | <tr> | ||
| + | <th style="background-color:#80BA6B ">Dilution</th> | ||
| + | <th style="background-color:#80BA6B ">PsDef1 (mg/mL) </th> | ||
| + | <th style="background-color:#80BA6B">SHuffle control (mg/mL) </th> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td>1 (undiluted)</td> | ||
| + | <td> 3.7402 </td> | ||
| + | <td> 3.74</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> 3:4</td> | ||
| + | <td> 2.8052</td> | ||
| + | <td>2.805 </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> 1:2 </td> | ||
| + | <td>1.8701</td> | ||
| + | <td>1.87</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> 1:4</td> | ||
| + | <td>0.9351</td> | ||
| + | <td> 0.935</td> | ||
| + | </tr> | ||
| + | </table> | ||
<br> | <br> | ||
| + | |||
| + | <p aling="justify"> | ||
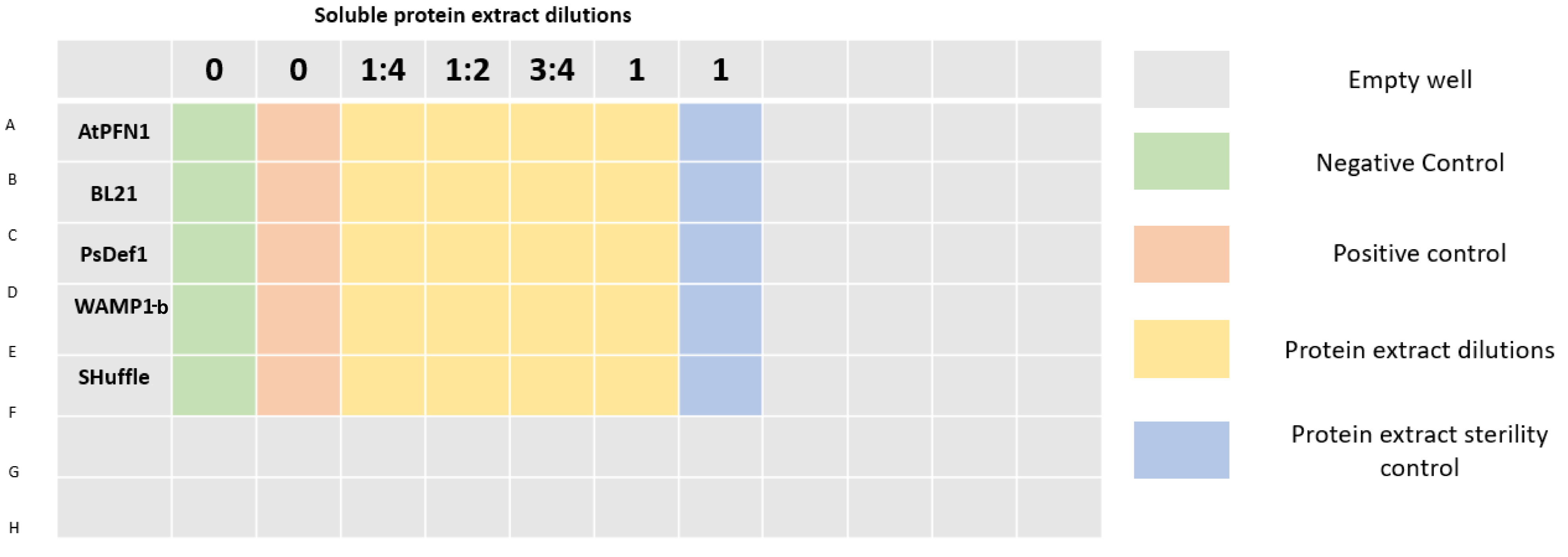
| + | 96 well plates were prepared as shown in Figure 3. A final volume of 200 μL was completed in each well by mixing protein extract, sterile distilled water (to achieve desired concentrations), potato dextrose broth, and a spore suspension of <i>V. dahliae</i> with a final concentration of 2x10<sup>4</sup> spores/mL per well. Plates were incubated at 25°C.</p> | ||
| + | <br> | ||
| + | [[File:T--Tec-Chihuahua--microFinal.png|800px]] | ||
| + | <center><b>Figure 3.</b> 96 well plate distribution.</center> | ||
| + | |||
<p align="justify"> | <p align="justify"> | ||
| − | + | Absorbance readings at 405 nm were performed in a Varioskan Lux 3020-231 microplate reader 1 and 24 hours after plate preparation. After analyzing the results, a growth rate percentage was estimated for every evaluated sample by using the following formula: | |
| + | </p> | ||
| + | <p align="justify"> | ||
| + | Growth rate = ((A<sub>1</sub> - A<sub>0</sub>)/A<sub>0</sub>) x 100 | ||
</p> | </p> | ||
| + | <p align="justify"> | ||
| + | Where:<br> | ||
| + | A<sub>1</sub> = Absorbance after 24 hours.<br> | ||
| + | A<sub>0</sub> = Absorbance after 1 hour. | ||
| + | </p> | ||
| + | |||
| + | <b>Results</b> | ||
<br> | <br> | ||
<p align="justify"> | <p align="justify"> | ||
| − | The | + | Results generated by the previous formula correspond to the percentage in which absorbance increased in each well compared to the initial reading. The results are summarized in figure 4. As the figure shows, an outstanding difference in growth values exists among the positive growth controls without any kind of protein extract, the untransformed strain extract controls, and the growth of the fungus with different peptide concentrations, was noted. The positive control was estimated to grow at a rate of 40.36% after 24 h, on the other hand, the lowest concentrations of added protein extracts clearly showed a decreasing behavior in growth rates. In comparison, growth rates from wells containing protein extracts of untransformed cells do not behave this way, meaning that PsDef1 had a degree of inhibition against <i>V. dahliae</i>, even in crude extract. |
| + | </p> | ||
| + | <p align="justify"> | ||
| + | In order to better comprehend the results, growth rates were used to estimate the percentage of inhibition using the following formula: | ||
</p> | </p> | ||
| − | + | <p align="justify"> | |
| − | <p align= "justify"> | + | I = 100 - (Gp/Gv) x 100<br> |
| − | + | Where:<br> | |
| + | I = Inhibition %<br> | ||
| + | Gp = Growth rate of <i>V. dahliae</i> treated with extract with peptides.<br> | ||
| + | Gv = Growth rate of <i>V. dahliae</i> positive control. | ||
</p> | </p> | ||
| − | |||
| − | |||
| − | <p | + | <p align="justify"> |
| − | + | During the evaluation of PsDef1, untransformed protein extracts seemed to increase the fungus’ growth in contrast with the extract containing the peptide. However, in figure 4, inhibition caused by PsDef1 can be observable in the two lowest dilutions, presenting high inhibition percentages of 82.23% and 70.81% for dilutions 1:2 and 1:4, respectively. These reductions in growth rate helped characterize the peptide’s reported antifungal activity. | |
</p> | </p> | ||
| + | [[File:T--Tec-Chihuahua--dos.jpeg|800px]] | ||
| + | <center><b>Figure 4.</b> Antifungal activity of PsDef1 against V. dahliae. V. dahliae growth rate (%) challenged with different concentrations of soluble protein extract from transformed E. coli SHuffle with PsDef1 and untransformed E. coli SHuffle.</center> | ||
| − | |||
| − | |||
| − | |||
| + | <b>Discussion</b> | ||
| + | <br> | ||
| + | <p align="justify"> | ||
| + | When evaluating the results, we looked for an explanation for the inhibitory effect of the peptide occuring at lower concentrations of crude extract. A common occurrence in microplates assays is the precipitation of the inhibitory components, which leads to inconsistent inhibition values.<sup>8</sup> Peptides tend to precipitate at certain concentrations, depending on their composition, solubility and even the buffer or medium they’re diluted in.<sup>8, 9</sup> Given that we have almost no concrete information about our peptides, and that we saw ourselves forced to use crude extract, precipitation at the highest concentrations we used is a feasible possibility. It has been remarked in literature that this incidence makes it almost impossible to determine the MIC (Minimal Inhibitory Concentration) through this method.<sup>8</sup> | ||
| + | </p> | ||
| + | |||
| + | <p align="justify"> | ||
| + | The precipitation of our peptides could have affected the optic density measurements causing variations that altered the overall results.<sup>9</sup> Or, most likely, the precipitation altered the inhibitory effect of our peptides at high concentrations. This is a common occurrence, and recommendations can be found through literature, research papers, and protocol manuals.<sup>10</sup> | ||
| + | </p> | ||
<!-- --> | <!-- --> | ||
| Line 50: | Line 137: | ||
===Sequence and Features=== | ===Sequence and Features=== | ||
<partinfo>BBa_K2959005 SequenceAndFeatures</partinfo> | <partinfo>BBa_K2959005 SequenceAndFeatures</partinfo> | ||
| − | + | <br> | |
| + | <b> | ||
===References=== | ===References=== | ||
| − | + | </b> | |
1. Khairutdinov, B. I., Ermakova, E. A., Yusypovych, Y. M., Bessolicina, E. K., Tarasova, N. B., Toporkova, Y. Y., ... & Nesmelova, I. V. (2017). NMR structure, conformational dynamics, and biological activity of PsDef1 defensin from Pinus sylvestris. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics, 1865(8), 1085-1094. doi:10.1016/j.bbapap.2017.05.012 | 1. Khairutdinov, B. I., Ermakova, E. A., Yusypovych, Y. M., Bessolicina, E. K., Tarasova, N. B., Toporkova, Y. Y., ... & Nesmelova, I. V. (2017). NMR structure, conformational dynamics, and biological activity of PsDef1 defensin from Pinus sylvestris. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics, 1865(8), 1085-1094. doi:10.1016/j.bbapap.2017.05.012 | ||
<br> | <br> | ||
| Line 63: | Line 151: | ||
5. Hatahet, F., Nguyen, V. D., Salo, K. E., & Ruddock, L. W. (2010). Disruption of reducing pathways is not essential for efficient disulfide bond formation in the cytoplasm of E. coli. Microbial cell factories, 9(1), 67. doi: 10.1186/1475-2859-9-67 | 5. Hatahet, F., Nguyen, V. D., Salo, K. E., & Ruddock, L. W. (2010). Disruption of reducing pathways is not essential for efficient disulfide bond formation in the cytoplasm of E. coli. Microbial cell factories, 9(1), 67. doi: 10.1186/1475-2859-9-67 | ||
<br> | <br> | ||
| − | 6. Lisowsky, T. (1996). Removal of an intron with unique 3′ branch site creates an amino‐terminal protein sequence directing the scERV1 gene product to mitochondria. Yeast, 12(15), 1501-1510. doi: 10.1002/(SICI)1097-0061(199612)12:15%3C1501::AID-YEA40%3E3.0.CO;2-H | + | 6. Lisowsky, T. (1996). Removal of an intron with unique 3′ branch site creates an amino‐terminal protein sequence directing the scERV1 gene product to mitochondria. Yeast, 12(15), 1501-1510. doi: 10.1002/(SICI)1097-0061(199612)12:15%3C1501::AID-YEA40%3E3.0.CO;2-H</p> |
| + | <br> | ||
| + | 7. Schwalbe, R., Steele-Moore, L., & Goodwin, A. C. (2007). Antimicrobial susceptibility testing protocols. Crc Press. | ||
| + | <br> | ||
| + | 8. Eloff, J. (1998). A Sensitive and Quick Microplate Method to Determine the Minimal Inhibitory Concentration of Plant Extracts for Bacteria. Planta Medica, 64(08), 711–713. doi:10.1055/s-2006-957563 | ||
| + | <br> | ||
| + | 9. Morishige, H., Mano, Y., Oguri, T., & Furuya, N. (2012). Comparison of four reading methods of broth microdilution based on the Clinical and Laboratory Standards Institute M27-A3 method for Candida spp. THE JAPANESE JOURNAL OF ANTIBIOTICS , 65(5), 335–347. Retrieved from http://jja-contents.wdc-jp.com/pdf/JJA65/65-5/65-5_335-347.pdf | ||
| + | <br> | ||
| + | 10. Berditsch, M. (2012). Two-fold Broth Microdilution Method for Determination of MIC . Institute for Bio and Geosciences. Retrieved from http://www.ibg.kit.edu/nmr/downloads/MICprotocoll_30_Jan2012.pdf | ||
<br> | <br> | ||
Latest revision as of 03:01, 22 October 2019
Expressible Pinus sylvestris Defensin 1 with Erv1p
This composite part consists of a lacI regulated promoter, ribosome binding site, a coding sequence for Pinus sylvestris Defensin 1 as a fusion protein with a 6x His-Tag, a second ribosome binding site, a coding sequence for Erv1p, and a double terminator. This construct allows the expression of PsDef1, an antifungal peptide from Scots pine seeds, in E. coli. Expression can be positively regulated by the addition of IPTG or lactose thanks to the lacl regulated promoter. The fusion of PsDef1 to a 6x His-Tag is intended for the purification of the peptide by immobilized metal affinity chromatography.
Since PsDef1 is a peptide with four disulfide bonds1, a modification hard to replicate on prokaryotic expression systems, the construct allows for the co-expression of the peptide with a truncated version of Erv1p. This is a protein from Saccharomyces cerevisiae capable of catalyzing the formation of disulfide bonds2.
Usage and Biology
PsDef1 is present in Pinus sylvestris seeds, vegetative and generative organs. The defensin encodes a protein of 83 amino acids in length, whose first 33 amino acids correspond to the ‘N-terminal signal peptide, it also has eight highly conserved cysteines residues that are predicted to form four disulfide bonds C3–C49, C14–C34, C20–C43, and C24–C451, which stabilize its structure making it resistant to environmental changes2. Its conformation consists in a cysteine-stabilized αβ-motif (CSαβ) with a prominent α-helix and a triple-stranded antiparallel β-sheet that is stabilized by the 4 disulfide bonds1.
Endogenous PsDef1 causes morphological changes in fungi mycelium when interacting with the sphingolipid membrane on fungal cell, disrupting the membrane integrity. Antifungal activity towards phytopathogenic fungi Botrytis cinerea, Fusarium oxysporum, Fusarium solani, and Heterobasidion annosum has been demonstrated; as well as its antimicrobial activity against Gram-positive (Bacillus pumilus) and Gram-negative bacteria (Pectobacterium carotovorum, Pseudomonas fluorescens)1.
For a successful expression in E. coli, it is indispensable to avoid interactions that result in the aggregation of folding intermediates. Disulfide bonds can be involved in the structural, catalytic and signaling roles of the protein, but the formation of disulfide bonds can have certain problems that can cause misfolding, aggregation and low yields during the production of the protein4. The usage of Erv1p has been proposed as a mechanism for the formation of disulfide bonds in recombinant proteins in prokaryotic expression systems. It has been shown that, thanks to its ability to form disulfide bonds de novo, co-expression of Erv1p allows the proper formation of disulfide bonded proteins in the cytoplasm of E. coli5.
The ERV1 gene from Saccharomyces cerevisiae encodes for a 189 amino acids protein, Erv1p, that is involved in different processes of mitochondrial biogenesis and maintenance2, 6. The protein shows a flavin-linked sulfhydryl oxidase enzymatic activity linked to the carboxy-terminal domain. Thus, Erv1p is capable of oxidizing thiol groups in proteins and catalyzing disulfide bond formation. A 15 kDa truncated version of Erv1p, consisting of the 117 amino acid residues carboxy-terminal domain, shows a similar or improved sulfhydryl oxidase activity compared to the full length protein2.
Characterization of Expressible Pinus sylvestris Defensin 1 with Erv1p
Our DNA sequence PsDef1-Erv1p was synthesized by IDT®️ with the Biobrick prefix and suffix flanking the composite part. This made possible the correct digestion with restriction enzymes EcoRI-HF and PstI. After the digestion, ligation was performed with T7 ligase in order to place our construct into the pSB1C3 linearized backbone with chloramphenicol resistance, which was previously digested with the same restriction enzymes. Using the SnapGene®️ software, we could model our ligated expression plasmid, and the final part resulted in a sequence of 3,106 bp. Thereupon, Escherichia coli SHuffle was transformed by heat shock for following antibiotic selection of clones.

The next step was to amplify our BioBrick sequence through colony PCR performed upon our transformed cells to confirm the presence of our expression plasmid inside of our chassis. With the help of the specific forward Biobrick prefix [BBa_G1004] and the specific reverse Biobrick suffix [BBa_G1005], we were able to amplify our sequence exclusively. Through an agarose gel we confirmed the correct transformation. The PCR action from SnapGene®️ was used to predict the size of the amplified sequences which resulted in a size of 1,095 for PsDef1+Erv1p.


Protein production
IPTG Induction and Extraction
Following the construction of the BioBrick, it was necessary to induce protein production. Production of PsDef1 was possible under Lacl promoter when induced with 0.2 or 0.4 mM IPTG at 30°C and 225 rpm. This was followed by protein extraction by lysis solution to which lysozyme was added in order to obtain our soluble peptides.
Antifungal Assay
In order to test the antifungal activity of our peptides as well as prove their viability as a mechanism to inhibit Verticillium dahliae, an antifungal susceptibility test on a 96 well plate was carried out by measuring absorbance at 405 nm, wavelength used in standardized protocols to measure growth of filamentous fungi7. Due to a lack of time, we couldn’t reach the experimental stage of the project of peptide purification, so experiments were made using soluble protein extracts from our transformed cells’ lysates. Different dilutions of the extracts were prepared which were applied to a spore suspension of V. dahliae. Dilutions of the extracts of untransformed cells were used as controls to prove that inhibition was the result of the peptides and not any other protein contained within the extract. Soluble proteins from E. coli SHuffle were used as control for the PsDef1 extract. Concentrations of extracts with peptides were equalized to their controls. Table 1 details the concentrations of each dilution.
| Dilution | PsDef1 (mg/mL) | SHuffle control (mg/mL) |
|---|---|---|
| 1 (undiluted) | 3.7402 | 3.74 |
| 3:4 | 2.8052 | 2.805 |
| 1:2 | 1.8701 | 1.87 |
| 1:4 | 0.9351 | 0.935 |
96 well plates were prepared as shown in Figure 3. A final volume of 200 μL was completed in each well by mixing protein extract, sterile distilled water (to achieve desired concentrations), potato dextrose broth, and a spore suspension of V. dahliae with a final concentration of 2x104 spores/mL per well. Plates were incubated at 25°C.
Absorbance readings at 405 nm were performed in a Varioskan Lux 3020-231 microplate reader 1 and 24 hours after plate preparation. After analyzing the results, a growth rate percentage was estimated for every evaluated sample by using the following formula:
Growth rate = ((A1 - A0)/A0) x 100
Where:
A1 = Absorbance after 24 hours.
A0 = Absorbance after 1 hour.
Results
Results generated by the previous formula correspond to the percentage in which absorbance increased in each well compared to the initial reading. The results are summarized in figure 4. As the figure shows, an outstanding difference in growth values exists among the positive growth controls without any kind of protein extract, the untransformed strain extract controls, and the growth of the fungus with different peptide concentrations, was noted. The positive control was estimated to grow at a rate of 40.36% after 24 h, on the other hand, the lowest concentrations of added protein extracts clearly showed a decreasing behavior in growth rates. In comparison, growth rates from wells containing protein extracts of untransformed cells do not behave this way, meaning that PsDef1 had a degree of inhibition against V. dahliae, even in crude extract.
In order to better comprehend the results, growth rates were used to estimate the percentage of inhibition using the following formula:
I = 100 - (Gp/Gv) x 100
Where:
I = Inhibition %
Gp = Growth rate of V. dahliae treated with extract with peptides.
Gv = Growth rate of V. dahliae positive control.
During the evaluation of PsDef1, untransformed protein extracts seemed to increase the fungus’ growth in contrast with the extract containing the peptide. However, in figure 4, inhibition caused by PsDef1 can be observable in the two lowest dilutions, presenting high inhibition percentages of 82.23% and 70.81% for dilutions 1:2 and 1:4, respectively. These reductions in growth rate helped characterize the peptide’s reported antifungal activity.
Discussion
When evaluating the results, we looked for an explanation for the inhibitory effect of the peptide occuring at lower concentrations of crude extract. A common occurrence in microplates assays is the precipitation of the inhibitory components, which leads to inconsistent inhibition values.8 Peptides tend to precipitate at certain concentrations, depending on their composition, solubility and even the buffer or medium they’re diluted in.8, 9 Given that we have almost no concrete information about our peptides, and that we saw ourselves forced to use crude extract, precipitation at the highest concentrations we used is a feasible possibility. It has been remarked in literature that this incidence makes it almost impossible to determine the MIC (Minimal Inhibitory Concentration) through this method.8
The precipitation of our peptides could have affected the optic density measurements causing variations that altered the overall results.9 Or, most likely, the precipitation altered the inhibitory effect of our peptides at high concentrations. This is a common occurrence, and recommendations can be found through literature, research papers, and protocol manuals.10
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 580
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
References
1. Khairutdinov, B. I., Ermakova, E. A., Yusypovych, Y. M., Bessolicina, E. K., Tarasova, N. B., Toporkova, Y. Y., ... & Nesmelova, I. V. (2017). NMR structure, conformational dynamics, and biological activity of PsDef1 defensin from Pinus sylvestris. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics, 1865(8), 1085-1094. doi:10.1016/j.bbapap.2017.05.012
2. Lee, J. E., Hofhaus, G., & Lisowsky, T. (2000). Erv1p from Saccharomyces cerevisiae is a FAD‐linked sulfhydryl oxidase. FEBS letters, 477(1-2), 62-66. doi: 10.1016/s0014-5793(00)01767-1
3. Kovalyova, V. A., Gout, I. T., Kyamova, R. G., Filonenko, V. V., & Gout, R. T. (2007). Cloning and analysis of defensin 1 cDNA from Scots pine. Biopolymers and Cell,23(5), 398-404. doi: http://dx.doi.org/10.7124/bc.000779
4. Veggiani, G., & de Marco, A. (2011). Improved quantitative and qualitative production of single-domain intrabodies mediated by the co-expression of Erv1p sulfhydryl oxidase. Protein expression and purification, 79(1), 111-114. doi: 10.1016/j.pep.2011.03.005
5. Hatahet, F., Nguyen, V. D., Salo, K. E., & Ruddock, L. W. (2010). Disruption of reducing pathways is not essential for efficient disulfide bond formation in the cytoplasm of E. coli. Microbial cell factories, 9(1), 67. doi: 10.1186/1475-2859-9-67
6. Lisowsky, T. (1996). Removal of an intron with unique 3′ branch site creates an amino‐terminal protein sequence directing the scERV1 gene product to mitochondria. Yeast, 12(15), 1501-1510. doi: 10.1002/(SICI)1097-0061(199612)12:15%3C1501::AID-YEA40%3E3.0.CO;2-H</p>
7. Schwalbe, R., Steele-Moore, L., & Goodwin, A. C. (2007). Antimicrobial susceptibility testing protocols. Crc Press.
8. Eloff, J. (1998). A Sensitive and Quick Microplate Method to Determine the Minimal Inhibitory Concentration of Plant Extracts for Bacteria. Planta Medica, 64(08), 711–713. doi:10.1055/s-2006-957563
9. Morishige, H., Mano, Y., Oguri, T., & Furuya, N. (2012). Comparison of four reading methods of broth microdilution based on the Clinical and Laboratory Standards Institute M27-A3 method for Candida spp. THE JAPANESE JOURNAL OF ANTIBIOTICS , 65(5), 335–347. Retrieved from http://jja-contents.wdc-jp.com/pdf/JJA65/65-5/65-5_335-347.pdf
10. Berditsch, M. (2012). Two-fold Broth Microdilution Method for Determination of MIC . Institute for Bio and Geosciences. Retrieved from http://www.ibg.kit.edu/nmr/downloads/MICprotocoll_30_Jan2012.pdf