Difference between revisions of "Part:BBa K081005:Experience"
| (5 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
__NOTOC__ | __NOTOC__ | ||
This experience page is provided so that any user may enter their experience using this part.<BR>Please enter | This experience page is provided so that any user may enter their experience using this part.<BR>Please enter | ||
| Line 15: | Line 14: | ||
<I>Username</I> | <I>Username</I> | ||
|width='60%' valign='top'| | |width='60%' valign='top'| | ||
| − | Enter the review | + | Enter the review information here. |
|}; | |}; | ||
<!-- End of the user review template --> | <!-- End of the user review template --> | ||
<!-- DON'T DELETE --><partinfo>BBa_K081005 EndReviews</partinfo> | <!-- DON'T DELETE --><partinfo>BBa_K081005 EndReviews</partinfo> | ||
| + | |||
| + | {|width='80%' style='border:1px solid gray' | ||
| + | |- | ||
| + | |width='10%'| | ||
| + | <partinfo>BBa_K081005 AddReview 1</partinfo> | ||
| + | <I>iGEM 2009 British_Columbia</I> | ||
| + | |width='60%' valign='top'| | ||
| + | The part, as received in the 2009 iGEM distribution, did not sequence properly and, when assembled as a prefix with coding sequences (e.g. <partinfo>E1010</partinfo>), did not result in reporter expression. | ||
| + | |} | ||
| + | |||
| + | In 2019, team Sao_Carlos-Brazil characterized this part in E. coli as it follows: | ||
| + | |||
| + | ===Expression test=== | ||
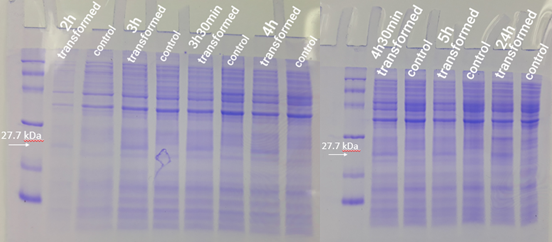
| + | 2.5 mL of transformed bacteria were inoculated into 250 mL of LB medium and incubated at 37°C at 120 rpm. Bacteria without the plasmid were also inoculated and used as a control group. Then, 500 mL aliquots were taken from each of the strains (transformed and not transformed) at 1h, 2h, 3h, 3h30min, 4h, 4h30min, 5h, and 24h, counted from the incubation. To follow the bacterial growth rate, the optical density was also inferred at each period. Aliquots removed were centrifuged for 1 minute at 16000g. Then, the formed pellet was resuspended in freshly prepared running buffer (800 µl Loading Buffer 4x, 200 µl DTT). For the first point (1h period), 30 µl of buffer was used for resuspension. For the other points, 60 µl of buffer was used. Then the mixture was boiled for 5 minutes. Test analysis was performed using 15% SDS PAGE gel, applying equalized sample volumes. | ||
| + | |||
| + | ===Results=== | ||
| + | |||
| + | The expression test presented positive results for the expression of the desired protein. In figure 1, it is possible to visualize bands of expected size (27.7 kDa) in the transformed strain samples, while in the control group there is no band of the mentioned size. This demonstrates that D. radiodurans Uracil DNA glycosylase protein was successfully expressed by the strong constitutive promoter associated with a strong RBS. This provided support for phenotypic testing, with irradiation of samples at progressive doses of ultraviolet C radiation. | ||
| + | |||
| + | <center> | ||
| + | https://2019.igem.org/wiki/images/8/89/T--Sao_Carlos-Brazil--Registry06.png | ||
| + | Figure 01</center> | ||
Latest revision as of 01:16, 22 October 2019
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_K081005
User Reviews
UNIQ7697e3d763563ab9-partinfo-00000000-QINU UNIQ7697e3d763563ab9-partinfo-00000001-QINU
|
•
iGEM 2009 British_Columbia |
The part, as received in the 2009 iGEM distribution, did not sequence properly and, when assembled as a prefix with coding sequences (e.g. BBa_E1010), did not result in reporter expression. |
In 2019, team Sao_Carlos-Brazil characterized this part in E. coli as it follows:
Expression test
2.5 mL of transformed bacteria were inoculated into 250 mL of LB medium and incubated at 37°C at 120 rpm. Bacteria without the plasmid were also inoculated and used as a control group. Then, 500 mL aliquots were taken from each of the strains (transformed and not transformed) at 1h, 2h, 3h, 3h30min, 4h, 4h30min, 5h, and 24h, counted from the incubation. To follow the bacterial growth rate, the optical density was also inferred at each period. Aliquots removed were centrifuged for 1 minute at 16000g. Then, the formed pellet was resuspended in freshly prepared running buffer (800 µl Loading Buffer 4x, 200 µl DTT). For the first point (1h period), 30 µl of buffer was used for resuspension. For the other points, 60 µl of buffer was used. Then the mixture was boiled for 5 minutes. Test analysis was performed using 15% SDS PAGE gel, applying equalized sample volumes.
Results
The expression test presented positive results for the expression of the desired protein. In figure 1, it is possible to visualize bands of expected size (27.7 kDa) in the transformed strain samples, while in the control group there is no band of the mentioned size. This demonstrates that D. radiodurans Uracil DNA glycosylase protein was successfully expressed by the strong constitutive promoter associated with a strong RBS. This provided support for phenotypic testing, with irradiation of samples at progressive doses of ultraviolet C radiation.