Difference between revisions of "Part:BBa K2559003"
(→NRP-UEA 2015) |
|||
| (17 intermediate revisions by 3 users not shown) | |||
| Line 16: | Line 16: | ||
The part is facilitate the in-depth research for other teams! | The part is facilitate the in-depth research for other teams! | ||
| − | == | + | == GZHS-United 2019 == |
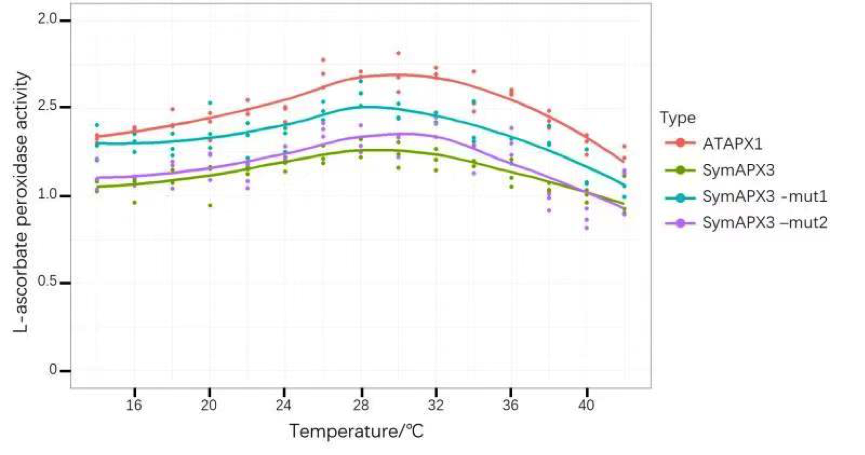
| − | + | We linked the gene of ATAPX1 to the existing promoter PrplJ, measured its enzyme activity in different states (different temperature, different illumination, different pH), and obtained correlation results between different factors and expressed enzyme protein activity. | |
| − | + | [[File:T--GZHS-United--Bronze-temperatures.png|800px|]] | |
| − | + | <i> Figure 1: This figure shows L-ascorbate peroxidase activity under different temperature, while ATAPX1 is control by PrplJ. </i> | |
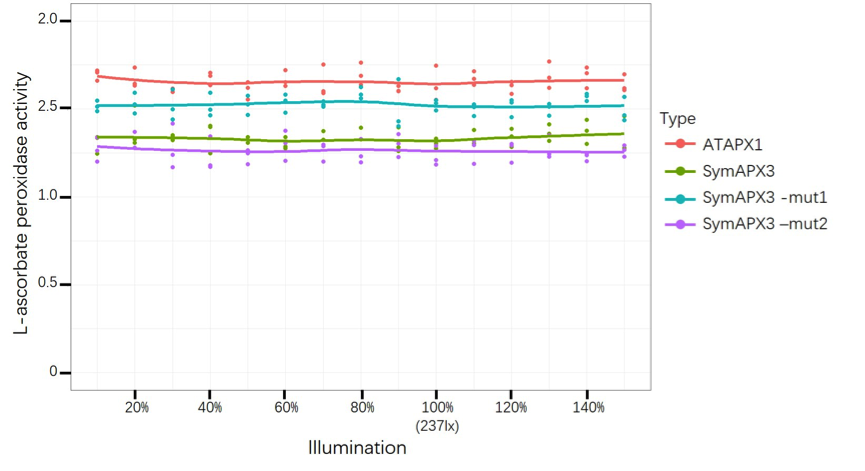
| + | [[File:T--GZHS-United--Bronze-illumination.png|800px|]] | ||
| − | + | <i> Figure 2: This figure shows L-ascorbate peroxidase activity under different illumination, while ATAPX1 is control by PrplJ. </i> | |
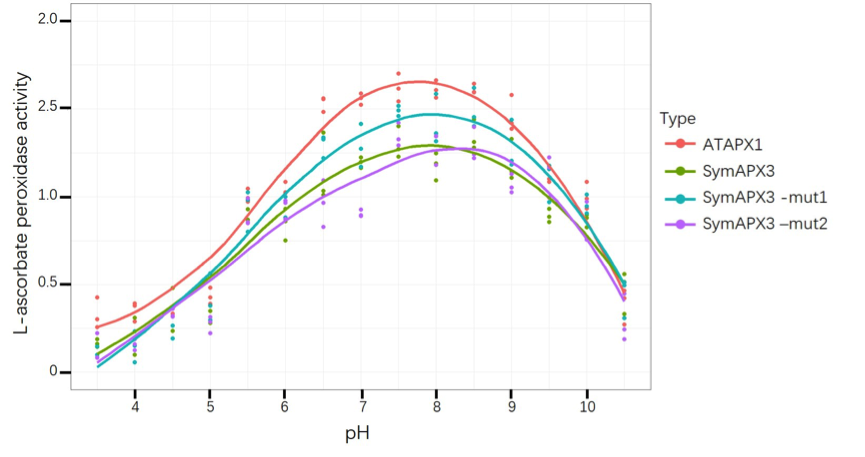
| − | <i> Figure | + | [[File:T--GZHS-United--Bronze-ph.png|800px|]] |
| + | |||
| + | <i> Figure 3: This figure shows L-ascorbate peroxidase activity under different pH, while ATAPX1 is control by PrplJ. </i> | ||
| + | |||
| + | Data which are obtained by our results, the above results indicate that we verify of the type of the promoter that PrplJ stability and reusability, especially in the experimental projects require different promoter can play a good role, such as the need to build contains multiple promoter and the need to use Gisbon assembly which is required to prevent the homologous recombination failure. When there is a must to use different promoter, PrplJ is a reliable choice. | ||
| + | |||
| + | You can see more details in our model page! https://2019.igem.org/Team:GZHS-United/Model | ||
| + | |||
| + | == JiangnanU_China 2019 == | ||
| + | |||
| + | In 2018, the team iGEM18_SCAU-China chose a strong E. coli endogenous promoter (PrplJ,BBa_K2559003) to express their amended eGFP (BBa_K2559005). This year our team JiangnanU_China also chose this promoter PrplJ (BBa_K2559003) to express their amended eGFP (BBa_K2559005). The team iGEM18_SCAU-China chose one timepoint to test the promoter strength. However, we measured the value of (fluorescence intensity)/OD600 at 0 h, 4 h, 6 h, 8 h and 10 h. Through the change of value what we measured, we can found the promoter PrplJ worked well. In the other hand, we observed the eGFP fluorescent protein by fluorescence microscope and we found that the E. coli with the amended eGFP fluorescent protein emitted the noticeable green fluorescence. | ||
| + | |||
| + | [[Image:PrplJ-eGFP-1.png|700px|thumb|center|Figure1. The curve of Fluorescent intensity/OD600 - Time(h)]] | ||
| + | |||
| + | [[Image:PrplJ-eGFP-2.png|650px|thumb|center|Figure2. Fluorescent intensity of eGFP driven by by PrplJ promoter]] | ||
| − | |||
<!-- --> | <!-- --> | ||
| − | + | == Sequence and Features == | |
<partinfo>BBa_K2559003 SequenceAndFeatures</partinfo> | <partinfo>BBa_K2559003 SequenceAndFeatures</partinfo> | ||
Latest revision as of 08:55, 19 October 2019
Ecoli promoter of sgRNA(PrplJ)
The BBa_K2559003 , PrplJ promoter is a strong endogenous promoter in Escherichia coli K 12. We used it to express single guide RNA in our CRISPR/cas9 system.
Usage and Biology
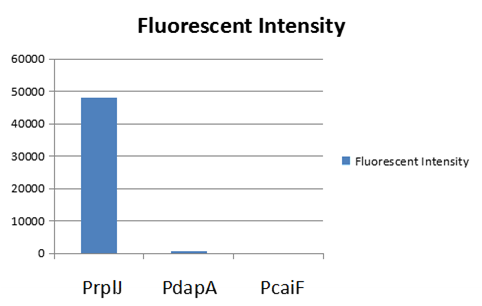
We obtained this promoter from a database named PromEC ( http://margalit.huji.ac.il/promec/index.html) .PromEC is an updated compilation of E. coli mRNA promoter sequences. It includes detailed information of genesites which have been experimentally identified mRNA transcriptional starting position in the E. coli chromosome, as well as the actual sequences of the promoters. We have built up a model to measure the expression intensity of these promoters. Eventually, we selected three promoters including: PrplJ, Pdapa and PcaiF based on the results of modeling PrplJ, Pdapa and PcaiF.We further made chimeric fluorescent fusion proteins between the promoter and GFP respectively and tested the fluorescent intensity driven by different promoters. The combination of PromEC and our modeling can be used as a efficient tool to figure out the information about interested promoters. The GFP expression in E.coli (DH5 α) and was driven by PrplJ, Pdapa and PcaiF promoter.
Basing on the model predition and experiment testing results,we used PrplJ as the promoter of sgRNA
The part is facilitate the in-depth research for other teams!
GZHS-United 2019
We linked the gene of ATAPX1 to the existing promoter PrplJ, measured its enzyme activity in different states (different temperature, different illumination, different pH), and obtained correlation results between different factors and expressed enzyme protein activity.
Figure 1: This figure shows L-ascorbate peroxidase activity under different temperature, while ATAPX1 is control by PrplJ.
Figure 2: This figure shows L-ascorbate peroxidase activity under different illumination, while ATAPX1 is control by PrplJ.
Figure 3: This figure shows L-ascorbate peroxidase activity under different pH, while ATAPX1 is control by PrplJ.
Data which are obtained by our results, the above results indicate that we verify of the type of the promoter that PrplJ stability and reusability, especially in the experimental projects require different promoter can play a good role, such as the need to build contains multiple promoter and the need to use Gisbon assembly which is required to prevent the homologous recombination failure. When there is a must to use different promoter, PrplJ is a reliable choice.
You can see more details in our model page! https://2019.igem.org/Team:GZHS-United/Model
JiangnanU_China 2019
In 2018, the team iGEM18_SCAU-China chose a strong E. coli endogenous promoter (PrplJ,BBa_K2559003) to express their amended eGFP (BBa_K2559005). This year our team JiangnanU_China also chose this promoter PrplJ (BBa_K2559003) to express their amended eGFP (BBa_K2559005). The team iGEM18_SCAU-China chose one timepoint to test the promoter strength. However, we measured the value of (fluorescence intensity)/OD600 at 0 h, 4 h, 6 h, 8 h and 10 h. Through the change of value what we measured, we can found the promoter PrplJ worked well. In the other hand, we observed the eGFP fluorescent protein by fluorescence microscope and we found that the E. coli with the amended eGFP fluorescent protein emitted the noticeable green fluorescence.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]