Difference between revisions of "Part:BBa K3187015"
PeterGockel (Talk | contribs) |
PeterGockel (Talk | contribs) |
||
| (3 intermediate revisions by the same user not shown) | |||
| Line 23: | Line 23: | ||
target="_blank"> BBa_B0032</a>). The improved and the original version of the part were cloned into the pSB1C3 backbone and transformed in <i>E. coli</i> BL21 (DE3). | target="_blank"> BBa_B0032</a>). The improved and the original version of the part were cloned into the pSB1C3 backbone and transformed in <i>E. coli</i> BL21 (DE3). | ||
</p> | </p> | ||
| − | + | The part was improved in terms of its expression level, based on the insertion of a 5’-untranslated region (5’-UTR) upstream of the coding sequence. This 5'-UTR was adapted from iGEM Bielefeld 2015 ( <a | |
href="https://parts.igem.org/Part:BBa_K1758100" | href="https://parts.igem.org/Part:BBa_K1758100" | ||
target="_blank"> BBa_K1758100</a>) and is based on the research of Olins <i>et</i> al | target="_blank"> BBa_K1758100</a>) and is based on the research of Olins <i>et</i> al | ||
| Line 117: | Line 117: | ||
<br></br> | <br></br> | ||
<p> | <p> | ||
| − | <b>Fig. | + | <b>Fig. 2</b> shows the original version of the part (left) and the improved version (right) after overnight cultivation on an agar plate and after cultivation for 24 h in M9 minimal medium for better visualization of the emitted light at 512 nm<sup id="cite_ref-5" class="reference"> |
<a href="#cite_note-5">[5] </a> </sup>. Both the agar plate and the liquid culture were supplemented with chloramphenicol according to the cmp resistance on the pSB1C3 backbone. Both pictures were taken using a dark reader (Dark Reader Clare Chemical Research) for better visualization of the yellow color. | <a href="#cite_note-5">[5] </a> </sup>. Both the agar plate and the liquid culture were supplemented with chloramphenicol according to the cmp resistance on the pSB1C3 backbone. Both pictures were taken using a dark reader (Dark Reader Clare Chemical Research) for better visualization of the yellow color. | ||
</p> | </p> | ||
| Line 138: | Line 138: | ||
Both the agar plate and the liquid culture show an enhanced expression of the amilGFP gene after the upstream insertion of the translation enhancing 5’-UTR. | Both the agar plate and the liquid culture show an enhanced expression of the amilGFP gene after the upstream insertion of the translation enhancing 5’-UTR. | ||
</p> | </p> | ||
| + | <h2>References</h2> | ||
| + | <ol class="references"> | ||
| + | <li id="cite_note-1"> | ||
| + | <span class="mw-cite-backlink"> | ||
| + | <a href="#cite_ref-1">↑</a> | ||
| + | </span> | ||
| + | <span class="reference-text"> | ||
| + | Peter O. Olins and Shaukat H. Rangwala, A Novel Sequence Element Derived from Bacteriophage T7 mRNA Acts as an Enhancer of Translation of the lacZ Gene in Escherichia coli, The Journal of Biological Chemistry, Vol. 264, No. 29, Issue of Ovtobet 15, pp. 16973-16976, 1989 | ||
| + | <a rel="nofollow" class="external autonumber" href=" https://www.ncbi.nlm.nih.gov/pubmed/2676996" | ||
| + | target="_blank">[1] </a> | ||
| + | </span> | ||
| + | </li> | ||
| + | |||
| + | <li id="cite_note-2"> | ||
| + | <span class="mw-cite-backlink"> | ||
| + | <a href="#cite_ref-2">↑</a> | ||
| + | </span> | ||
| + | <span class="reference-text"> | ||
| + | Shuntaor Takahashi, Hiroyuki Furusawa, Takuya Ueda and Yoshio Okahata, Translation Enhancer Improves the Ribosome Liberation from Translation Initiation, J. Am. Chem. Soc. 2013, 135 35, 13096-13106 | ||
| + | <a rel="nofollow" class="external autonumber" | ||
| + | href=" https://www.ncbi.nlm.nih.gov/pubmed/23927491 | ||
| + | " target="_blank">[2] | ||
| + | </a> | ||
| + | </span> | ||
| + | </li> | ||
| + | |||
| + | <li id="cite_note-3"> | ||
| + | <span class="mw-cite-backlink"> | ||
| + | <a href="#cite_ref-3">↑</a> | ||
| + | </span> | ||
| + | <span class="reference-text"> | ||
| + | David K. Karig, Sukanya Iyer, Michael L. Simpson and Mitchel J. Doktycz, Expression optimization and synthetic gene networks in cell-free systems, Nucleic Acids Res. 2012 Apr; 40(8): 3763.3774 | ||
| + | <a rel="nofollow" class="external autonumber" | ||
| + | href="https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3333853/" target="_blank">[3] | ||
| + | </a> | ||
| + | </span> | ||
| + | </li> | ||
| + | |||
| + | <li id="cite_note-4"> | ||
| + | <span class="mw-cite-backlink"> | ||
| + | <a href="#cite_ref-4">↑</a> | ||
| + | </span> | ||
| + | <span class="reference-text"> | ||
| + | Roberta Lentini, Silvia Perez Santero, Fabio Chizzolini, Dario Cecchi, Jason Fontana, Marta Marchioretto, Christina Del Bianco, Jessica L. Terrell, Amy C. Spencer, Laura Martini, Michele Forlin, Michael Assfalg, Mauro Dalla Serra, William E. Bentley and Sheref S. Mansy, Integrating artificial with natural cells to translate chemical messages that direct E. coli behavior, Nature Communications 5, Article number: 4012 (2014) | ||
| + | <a rel="nofollow" class="external autonumber" | ||
| + | href=" https://www.nature.com/articles/ncomms5012 " | ||
| + | target="_blank">[4] </a> | ||
| + | </span> | ||
| + | </li> | ||
| + | |||
| + | <li id="cite_note-5"> | ||
| + | <span class="mw-cite-backlink"> | ||
| + | <a href="#cite_ref-5">↑</a> | ||
| + | </span> | ||
| + | <span class="reference-text"> | ||
| + | Joesfine Liljeruhm, Saskia K. Funk, Sandra Tietscher, Anders D. Edlund, Sabri Jamal, Pikkei Wistrand-Yuen, Karl Dyrhage, Arvid Gynna, Katarina Ivermark, Jessica Lövgren, Viktor Törnblom, Anders Virtanen, Erik R. Lundin, Erik Wistrand-Yuen and Anthony C. Forster, Enginerring a palette of eukaryotic chromoproteins for bacterial synthetic biology, Journal of Biological Engineering 12, Article number: 8 (2018) | ||
| + | |||
| + | <a rel="nofollow" class="external autonumber" | ||
| + | href=" https://jbioleng.biomedcentral.com/articles/10.1186/s13036-018-0100-0 " | ||
| + | target="_blank">[5] </a> | ||
| + | </span> | ||
| + | </li> | ||
| + | |||
| + | |||
| + | |||
| + | </ol> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
</html> | </html> | ||
Latest revision as of 19:58, 18 October 2019
Constitutively expressed chromoprotein amilGFP with Translation Enhancing 5'-UTR
Improved Version of BBa_K1073024
Usage and Biology
BBa_K3187015 (improved BBa_K1073024) encodes the yellow chromoprotein amilGFP derived from the coral Acropora millepora [5] under the control of a strong constitutive promoter (BBa_J23100) in combination with a ribosomal binding site ( BBa_B0032). The improved and the original version of the part were cloned into the pSB1C3 backbone and transformed in E. coli BL21 (DE3).
The part was improved in terms of its expression level, based on the insertion of a 5’-untranslated region (5’-UTR) upstream of the coding sequence. This 5'-UTR was adapted from iGEM Bielefeld 2015 ( BBa_K1758100) and is based on the research of Olins et al [1] and Takahashi et al. [2] . It contains the strong ribosomal binding site (RBS) g10-L from the T7 bacteriophage and a sequence that plays a role in the regulation of mRNA binding to and release from the 30S ribosomal subunit. The 5'-UTR therefore enhances the translation efficiency of the following coding sequence (CDS) (Fig. 1).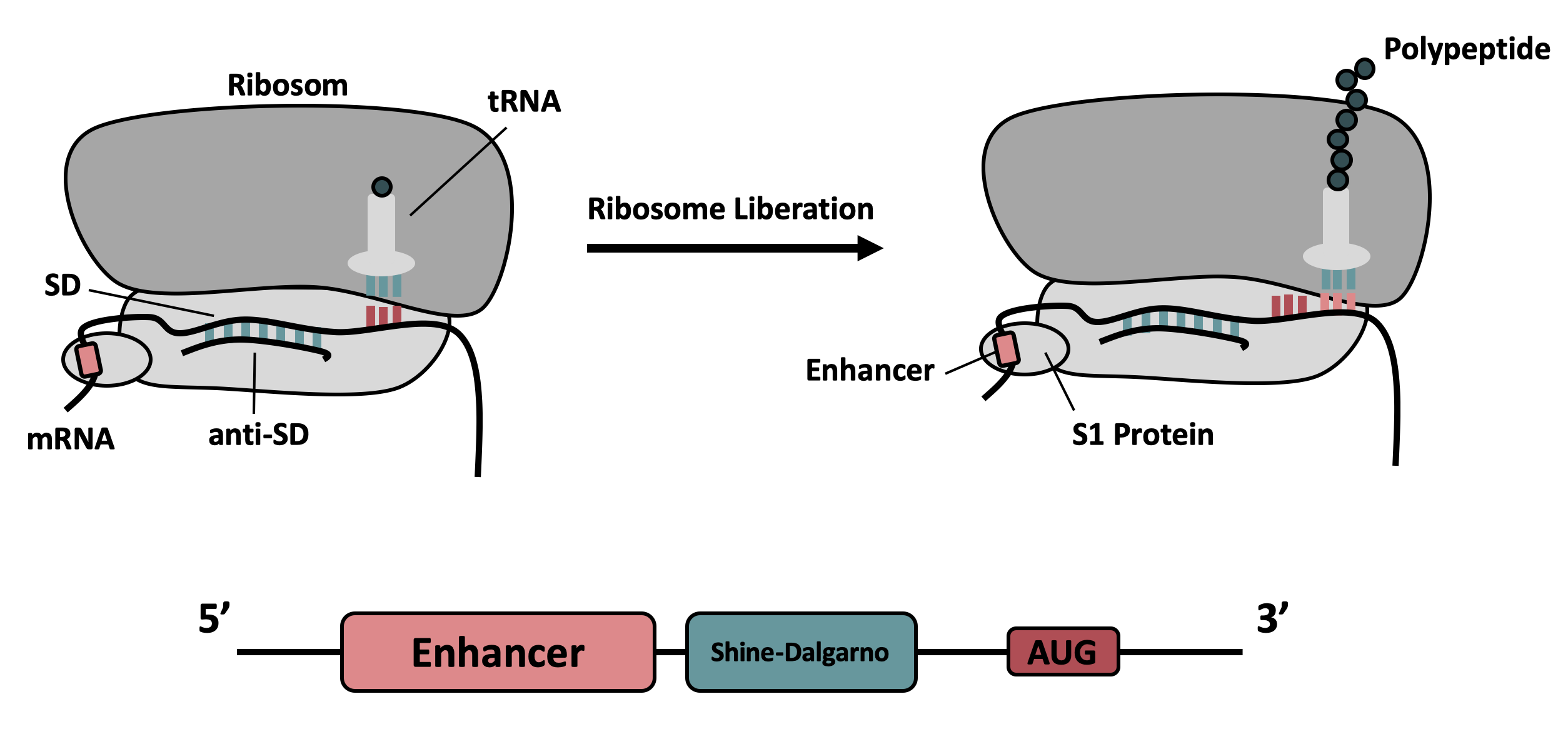
The sequence of the translation enhancing 5’-UTR can be divided into the four main features listed below:

| Sequence | Function |
|---|---|
|
AATAATTTTGTT TTAACTTTAA |
The T7 g10 leader sequence (first described by Olins et al [1] )increases the efficiency of translation initiation. This sequence contains the epsilon motif TTAACTTTA which enhances the binding of the mRNA to the 16S rRNA. |
| poly-A | Referring to Takahashi et al. [2] a spacer between the epsilon motive and the RBS improves the translation rate. |
| GAAGGAG | According to Karig et al. [3] and Lentini et. al [4] a distance of 4-9 bases between RBS and start codon increases the translation efficiency. |
| AATAATCT | According to Lentini et. al [4] an AT-rich composition between the RBS and the start codon results in the best expression results. |
Results
Fig. 2 shows the original version of the part (left) and the improved version (right) after overnight cultivation on an agar plate and after cultivation for 24 h in M9 minimal medium for better visualization of the emitted light at 512 nm [5] . Both the agar plate and the liquid culture were supplemented with chloramphenicol according to the cmp resistance on the pSB1C3 backbone. Both pictures were taken using a dark reader (Dark Reader Clare Chemical Research) for better visualization of the yellow color.
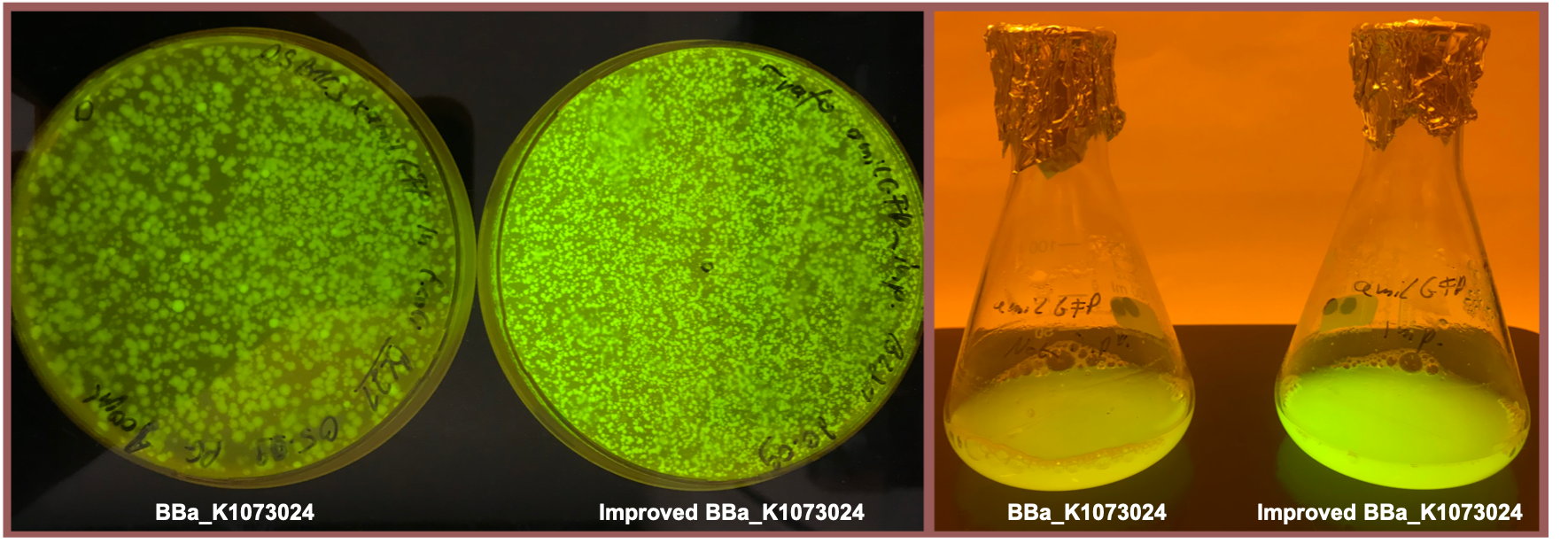
Both the agar plate and the liquid culture show an enhanced expression of the amilGFP gene after the upstream insertion of the translation enhancing 5’-UTR.
References
- ↑ Peter O. Olins and Shaukat H. Rangwala, A Novel Sequence Element Derived from Bacteriophage T7 mRNA Acts as an Enhancer of Translation of the lacZ Gene in Escherichia coli, The Journal of Biological Chemistry, Vol. 264, No. 29, Issue of Ovtobet 15, pp. 16973-16976, 1989 [1]
- ↑ Shuntaor Takahashi, Hiroyuki Furusawa, Takuya Ueda and Yoshio Okahata, Translation Enhancer Improves the Ribosome Liberation from Translation Initiation, J. Am. Chem. Soc. 2013, 135 35, 13096-13106 [2]
- ↑ David K. Karig, Sukanya Iyer, Michael L. Simpson and Mitchel J. Doktycz, Expression optimization and synthetic gene networks in cell-free systems, Nucleic Acids Res. 2012 Apr; 40(8): 3763.3774 [3]
- ↑ Roberta Lentini, Silvia Perez Santero, Fabio Chizzolini, Dario Cecchi, Jason Fontana, Marta Marchioretto, Christina Del Bianco, Jessica L. Terrell, Amy C. Spencer, Laura Martini, Michele Forlin, Michael Assfalg, Mauro Dalla Serra, William E. Bentley and Sheref S. Mansy, Integrating artificial with natural cells to translate chemical messages that direct E. coli behavior, Nature Communications 5, Article number: 4012 (2014) [4]
- ↑ Joesfine Liljeruhm, Saskia K. Funk, Sandra Tietscher, Anders D. Edlund, Sabri Jamal, Pikkei Wistrand-Yuen, Karl Dyrhage, Arvid Gynna, Katarina Ivermark, Jessica Lövgren, Viktor Törnblom, Anders Virtanen, Erik R. Lundin, Erik Wistrand-Yuen and Anthony C. Forster, Enginerring a palette of eukaryotic chromoproteins for bacterial synthetic biology, Journal of Biological Engineering 12, Article number: 8 (2018) [5]
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
