Difference between revisions of "Part:BBa K3017003"
| (19 intermediate revisions by 2 users not shown) | |||
| Line 5: | Line 5: | ||
[[File:T--Hong Kong HKUST--assgDNA_GFP_white.jpeg|thumb|Secondary structure of the transcription product of this part, predicted by NUPACK]] | [[File:T--Hong Kong HKUST--assgDNA_GFP_white.jpeg|thumb|Secondary structure of the transcription product of this part, predicted by NUPACK]] | ||
| − | |||
| − | |||
| + | <p>asRNA is the key to the reversibility of the CRISPRi switch. The asRNA binds to the artificial linker loop of the sgRNA, and extend the loop to become a linear structure. It is proven that this linearization of the loop causes derepression[1].</p> | ||
| + | <p>The synthetic asRNA is composed of 2 functional domains, extensor, for linearizing the loop, and Spot 42, aiding the stability of the asRNA.</p> | ||
| Line 15: | Line 15: | ||
<h2>Extensor</h2> | <h2>Extensor</h2> | ||
| − | <p>The extensor sequence is complementary to the artificial linker loop. By forming a duplex, the loop is extended. It was thought in the study that an asRNA complementary to the spacer would displace the DNA target and yield a higher depression effect. But after optimization, the study found out that changing the sgRNA’s secondary structure by extending a loop formed by the artificial linker would maximize the derepression effect to at least 95%, compared to only 15-55% of complementing the spacer.</p> | + | <p>The extensor sequence is complementary to the artificial linker loop. By forming a duplex, the loop is extended. It was originally thought in the study[1] that an asRNA complementary to the spacer would displace the DNA target and yield a higher depression effect. But after optimization, the study found out that changing the sgRNA’s secondary structure by extending a loop formed by the artificial linker would maximize the derepression effect to at least 95%, compared to only 15-55% of complementing the spacer[1].</p> |
| − | <p>The derepression is specific to the <i>gfp</i> gene BBa_E0040. Therefore asRNA extensor in the transcription product of this part targets sgRNA artificial linker of transcription product of part | + | <p>The derepression is specific to the <i>gfp</i> gene BBa_E0040. Therefore asRNA extensor in the transcription product of this part targets sgRNA artificial linker of transcription product of part BBa_K3017001.</p> |
| − | [[File:T--Hong Kong HKUST--spot42.png|thumb|Complex formation between asRNA, sgRNA and target DNA is depicted]] | + | [[File:T--Hong Kong HKUST--spot42.png|180px|thumb|Complex formation between asRNA, sgRNA and target DNA is depicted]] |
<h2>Spot 42</h2> | <h2>Spot 42</h2> | ||
| − | <p>Bacterial Hfq protein is known to modulate the stability or the translation of mRNAs and interact with small regulatory RNAs. A study provides evidence that Hfq strongly cooperates in intermolecular base pairing between the asRNA and its target RNA. Spot 42 is an RNA antisense regulator that has several A/U-rich regions that provide specificity to Hfq protein. Hfq protein also provide protection to the asRNA at these A/U-rich regions.</p> | + | <p>Bacterial Hfq protein is known to modulate the stability or the translation of mRNAs and interact with small regulatory RNAs. A study provides evidence that Hfq strongly cooperates in intermolecular base pairing between the asRNA and its target RNA. Spot 42 is an RNA antisense regulator that has several A/U-rich regions that provide specificity to Hfq protein. Hfq protein also provide protection to the asRNA at these A/U-rich regions.[5]</p> |
| + | |||
| + | <p>Same Spot 42 sequence is present in every asRNA we have designed as it is highly specific to Hfq protein and the duplex is necessary for the asRNA to work. Hfq is endogenous to <i>E.coli</i>, therefore the protein is not encoded in any part of the circuit[6].</p> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <h1>Characterizations</h1> | ||
| + | This part involve interaction with part BBa_K3017001 so they were being characterized as a pair. | ||
| + | Using sgRNA-terminator BBa_K3017061 and asRNA-terminator-BBa_K3017060 as a model, we have characterized this part which is highly similar in function. | ||
| + | |||
| + | <h2>RNA In Vitro Transcription (IVT) and interaction assay</h2> | ||
| + | |||
| + | <h4>Aim:</h4> | ||
| + | <p>To assess whether the sgRNA and asRNA can be transcribed successfully and hybridized <i>in vivo</i>, an <i>in vitro</i> assay was designed to assess the transcription of the RNAs and the feasibility for the RNAs to hybridize <i>in vitro</i>. | ||
| + | </p> | ||
| + | |||
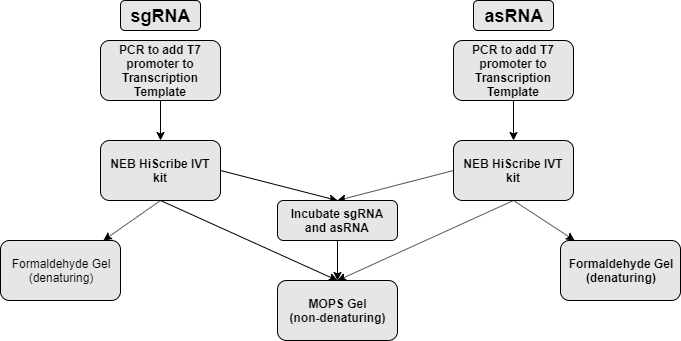
| + | <h4> Flow of experiment:</h4> | ||
| + | <p> | ||
| + | sgRNA and asRNA is synthesized via <i>in vitro</i> transcription (NEB HiScribe T7 <i>in vitro</i> transcription kit). Part of the transcript is mixed together to allow for hybridization. The part is then analyzed on both formaldehyde denaturing agarose gel and non-denaturing agarose gel to assess transcript size and hybridization respectively. (figure 1) | ||
| + | </p> | ||
| + | |||
| + | {| class="wikitable" | ||
| + | |- | ||
| + | |<b>Denaturing Gel (formaldehyde)</b> | ||
| + | |<b>Non-denaturing Gel</b> | ||
| + | |- | ||
| + | |Destroys secondary structure | ||
| + | |Preserves secondary structure | ||
| + | |- | ||
| + | |Assess actual size of transcript | ||
| + | |Assess interaction | ||
| + | |} | ||
| + | <br> | ||
| + | [[File:T--Hong_Kong_HKUST--RNAoverview.png|700px|thumb|<b>Figure 1.</b> Flow of RNA interaction assay|center]] | ||
| + | <br> | ||
| + | |||
| + | <h4>Step 1 - PCR synthesis of IVT templates</h4> | ||
| + | <p>The 2 RNA’s transcription templates, designed according to [https://international.neb.com/protocols/2015/11/24/sgrna-synthesis-using-the-hiscribe-quick-t7-high-yield-rna-synthesis-kit-neb-e2050 NEB IVT kit manual], were created by using PCR to insert a T7 promoter in front of the transcription template using an overhang forward primer. (Figure 2) | ||
| + | </p> | ||
| + | |||
| + | <br> | ||
| + | [[File:T--Hong_Kong_HKUST--RNA_IVTprimer1.png|700px|thumb|<b>Figure 2.</b> The RNA’s transcription templates|center]] | ||
| + | <br> | ||
| + | |||
| + | <p>Thereafter, the PCR product is run through an agarose gel to separate impurities and inspect the sizes of the IVT templates before undergoing gel purification.</p> | ||
| + | |||
| + | <h3> Results:</h3> | ||
| + | https://2019.igem.org/wiki/images/9/98/T--Hong_Kong_HKUST--Expfig3.png | ||
| + | <br><b>Figure 3.</b> Verification of RNA’s transcription templates | ||
| + | <br> | ||
| + | <p>The size of IVT template is as expected to be around 150bp, with sgRNA expected to be 155bp and asRNA expected to be 157bp. This proves the IVT template sizes are correct and can be purified for IVT reaction. (Figure 3) | ||
| + | </p> | ||
| + | <br> | ||
| + | |||
| + | <h3>Step 2 - <i>In vitro</i> transcription (IVT) using NEB T7 HiScribe Transcription Kit</h3> | ||
| + | <p> | ||
| + | To synthesize the sgRNA and asRNA, 75ng of gel-purified DNA transcription template was added into the IVT reaction mixture. The reaction is set for 16 hours of incubation at 37°C, using a thermal cycler to prevent evaporation. | ||
| + | <br><br> | ||
| + | The transcripted RNAs is <b>[https://international.neb.com/products/m0303-dnase-i-rnase-free#Product%20Information treated with DNase1]</b>, then precipitated by sodium acetate/ethanol. | ||
| + | <br><br> | ||
| + | The sizes of the 2 precipitated RNAs are confirmed by using denaturing formaldehyde-MOPS 2% agarose gel. Since sgRNA is known to form multiple secondary structures, heat denaturation at 65 °C for 7 minutes is needed prior to loading into the gel to remove all secondary structures, allowing for accurate assessment of its size. | ||
| + | </p> | ||
| + | |||
| + | <h3>Results:</h3> | ||
| + | https://2019.igem.org/wiki/images/d/de/T--Hong_Kong_HKUST--Expfig4.png | ||
| + | <br><b>Figure 4.</b> RNA transcription check | ||
| + | <br> | ||
| + | <p>The gel photo above clearly indicates the successful transcription of the sgRNA and asRNA, with the size matching their expected size (sgRNA=138bp, asRNA=140bp). | ||
| + | </p> | ||
| + | <br> | ||
| + | |||
| + | <h3>Step 3 - RNA <i>in vitro</i> hybridization and visualization</h3> | ||
| + | <p> | ||
| + | In order to test the environment needed for <i>in vitro</i> hybridization, 4 different environments were tested in a combination of using 3x Hybridization Buffer (HB) and the use of initial denaturation. Regardless of the conditions being tested, the 2 RNAs are incubated together at 37°C for 45 minutes, before snap cooling to 0°C to preserve the hybridized structure and RNase degradation. ([https://2019.igem.org/wiki/images/6/69/T--Hong_Kong_HKUST--RNAannealingprot.pdf protocol we used for the experiment]) | ||
| + | <br><br> | ||
| + | It is known that salt content is crucial to RNA duplex formation, thus, 3x HB is used to provide the necessary salt content and stabilize the RNAs. DEPC water is used as a negative control to the testing of hybridization buffer. | ||
| + | <br><br> | ||
| + | Since this assay is designed as an <i>in vitro</i> demonstration of the <i>in vivo</i> hybridization, the temperature of incubation is set to 37°C to simulate the <i>in vivo</i> environment with no initial denaturation. As a fail-safe test, the necessity for initial denaturation is also tested as the <i>in vitro</i> hybridization is expected to be less efficient without the Hfq Chaperone. Therefore, denaturing the RNAs before slow cooling will remove the RNAs secondary structure while promoting the complementation and hybridization during slow cooling. | ||
| + | <br> | ||
| + | A non-denaturing 2% MOPS agarose gel is then used to visualize the hybridization. | ||
| + | </p> | ||
| + | |||
| + | <br> | ||
| + | <h3>Results:</h3> | ||
| + | https://2019.igem.org/wiki/images/3/30/T--Hong_Kong_HKUST--Expfig5.png | ||
| + | <br><b>Figure 5. </b>RNA hybridization assay | ||
| + | |||
| + | {| class="wikitable" | ||
| + | |+Index | ||
| + | |- | ||
| + | |HB/D | ||
| + | |Hybridization Buffer + denature + slow cool* + 37°C incubation | ||
| + | |- | ||
| + | |HB/37 | ||
| + | |Hybridization Buffer + slow cool* + 37°C incubation | ||
| + | |- | ||
| + | |D/D | ||
| + | |DEPC H2O + denature + slow cool* +37°C incubation | ||
| + | |- | ||
| + | |D/37 | ||
| + | |DEPC H2O + 37°C incubation | ||
| + | |- | ||
| + | |L | ||
| + | |Ambion Century™ - Plus RNA Markers | ||
| + | |- | ||
| + | |sg | ||
| + | |sgRNA BBa_K3017061 (138bp) | ||
| + | |- | ||
| + | |as | ||
| + | |asRNA BBa_K3017060 (140bp) | ||
| + | |} | ||
| + | <br>*slow cool is done by cooling the RNA sample using thermo cycler at 5°C per 10 sec until incubation temperature | ||
| − | <p> | + | <p> |
| + | As shown in the above gel photo (Figure 5), only lane AB/37 shows an extra band which appears to be around 250bp when compared to the ladder. This band indicates the hybridization of sgRNA and asRNA and is therefore running slower in agarose gel. | ||
| + | <br><br> | ||
| + | This result shows that salt content provided by the hybridization buffer (60mM KCl, 6mM HEPES, 0.2mM MgCl2 at 1x), which is similar to that of a cellular salinity, is crucial to the hybridization. This result also indicates that the RNAs are able to hybridize with each other in 37°C incubation with no initial denaturation needed. These results suggest that the RNAs are capable of hybridizing <i>in vivo</i>. With endogenous expression of Hfq protein in <i>E.coli</i>, the asRNA will also stabilize due to Spot 42 and attract other ssRNAs to hybridize. | ||
| + | <br><br> | ||
| + | Thus, we suspect the <i>in vivo</i> hybridization efficiency would be higher than our results. | ||
| + | </p> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <p><b>References:</b></p> | ||
| + | <br>[1] Y. J. Lee, A. Hoynes-Oconnor, M. C. Leong, and T. S. Moon, “Programmable control of bacterial gene expression with the combined CRISPR and antisense RNA system,” Nucleic Acids Research, vol. 44, no. 5, pp. 2462–2473, Feb. 2016. | ||
| + | <br>[2] C. Anders, O. Niewoehner, A. Duerst, and M. Jinek, “Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease,” Nature, vol. 513, no. 7519, pp. 569–573, 2014. | ||
| + | <br>[3] S. H. Sternberg, S. Redding, M. Jinek, E. C. Greene, and J. A. Doudna, “DNA Interrogation by the CRISPR RNA-Guided Endonuclease Cas9,” Biophysical Journal, vol. 106, no. 2, 2014. | ||
| + | <br>[4] T. Karvelis, G. Gasiunas, A. Miksys, R. Barrangou, P. Horvath, and V. Siksnys, “crRNA and tracrRNA guide Cas9-mediated DNA interference inStreptococcus thermophilus,” RNA Biology, vol. 10, no. 5, pp. 841–851, 2013. | ||
| + | <br>[5] T. Møller, T. Franch, P. Højrup, D. R. Keene, H. P. Bächinger, R. G. Brennan, and P. Valentin-Hansen, “Hfq,” Molecular Cell, vol. 9, no. 1, pp. 23–30, 2002. | ||
| + | <br>[6] G. M. Cech, A. Szalewska-Pałasz, K. Kubiak, A. Malabirade, W. Grange, V. Arluison, and G. Węgrzyn, “The Escherichia Coli Hfq Protein: An Unattended DNA-Transactions Regulator,” Frontiers in Molecular Biosciences, vol. 3, 2016. | ||
Latest revision as of 01:36, 22 October 2019
CRISPRi antisense RNA for derepressing gfp - transcription template
asRNA is the key to the reversibility of the CRISPRi switch. The asRNA binds to the artificial linker loop of the sgRNA, and extend the loop to become a linear structure. It is proven that this linearization of the loop causes derepression[1].
The synthetic asRNA is composed of 2 functional domains, extensor, for linearizing the loop, and Spot 42, aiding the stability of the asRNA.
Extensor
The extensor sequence is complementary to the artificial linker loop. By forming a duplex, the loop is extended. It was originally thought in the study[1] that an asRNA complementary to the spacer would displace the DNA target and yield a higher depression effect. But after optimization, the study found out that changing the sgRNA’s secondary structure by extending a loop formed by the artificial linker would maximize the derepression effect to at least 95%, compared to only 15-55% of complementing the spacer[1].
The derepression is specific to the gfp gene BBa_E0040. Therefore asRNA extensor in the transcription product of this part targets sgRNA artificial linker of transcription product of part BBa_K3017001.
Spot 42
Bacterial Hfq protein is known to modulate the stability or the translation of mRNAs and interact with small regulatory RNAs. A study provides evidence that Hfq strongly cooperates in intermolecular base pairing between the asRNA and its target RNA. Spot 42 is an RNA antisense regulator that has several A/U-rich regions that provide specificity to Hfq protein. Hfq protein also provide protection to the asRNA at these A/U-rich regions.[5]
Same Spot 42 sequence is present in every asRNA we have designed as it is highly specific to Hfq protein and the duplex is necessary for the asRNA to work. Hfq is endogenous to E.coli, therefore the protein is not encoded in any part of the circuit[6].
Characterizations
This part involve interaction with part BBa_K3017001 so they were being characterized as a pair. Using sgRNA-terminator BBa_K3017061 and asRNA-terminator-BBa_K3017060 as a model, we have characterized this part which is highly similar in function.
RNA In Vitro Transcription (IVT) and interaction assay
Aim:
To assess whether the sgRNA and asRNA can be transcribed successfully and hybridized in vivo, an in vitro assay was designed to assess the transcription of the RNAs and the feasibility for the RNAs to hybridize in vitro.
Flow of experiment:
sgRNA and asRNA is synthesized via in vitro transcription (NEB HiScribe T7 in vitro transcription kit). Part of the transcript is mixed together to allow for hybridization. The part is then analyzed on both formaldehyde denaturing agarose gel and non-denaturing agarose gel to assess transcript size and hybridization respectively. (figure 1)
| Denaturing Gel (formaldehyde) | Non-denaturing Gel |
| Destroys secondary structure | Preserves secondary structure |
| Assess actual size of transcript | Assess interaction |
Step 1 - PCR synthesis of IVT templates
The 2 RNA’s transcription templates, designed according to NEB IVT kit manual, were created by using PCR to insert a T7 promoter in front of the transcription template using an overhang forward primer. (Figure 2)
Thereafter, the PCR product is run through an agarose gel to separate impurities and inspect the sizes of the IVT templates before undergoing gel purification.
Results:
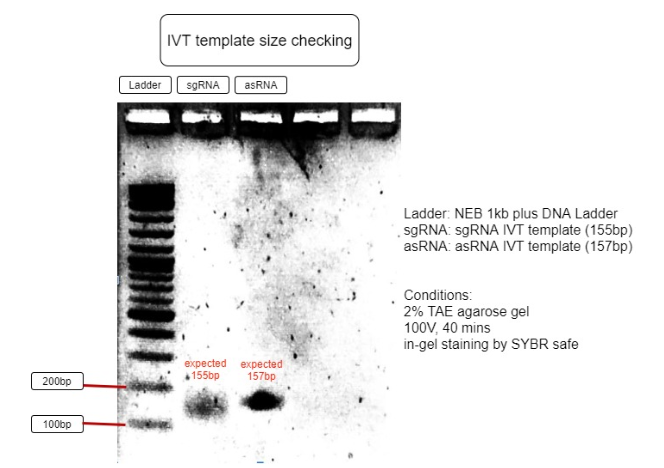
Figure 3. Verification of RNA’s transcription templates
The size of IVT template is as expected to be around 150bp, with sgRNA expected to be 155bp and asRNA expected to be 157bp. This proves the IVT template sizes are correct and can be purified for IVT reaction. (Figure 3)
Step 2 - In vitro transcription (IVT) using NEB T7 HiScribe Transcription Kit
To synthesize the sgRNA and asRNA, 75ng of gel-purified DNA transcription template was added into the IVT reaction mixture. The reaction is set for 16 hours of incubation at 37°C, using a thermal cycler to prevent evaporation.
The transcripted RNAs is treated with DNase1, then precipitated by sodium acetate/ethanol.
The sizes of the 2 precipitated RNAs are confirmed by using denaturing formaldehyde-MOPS 2% agarose gel. Since sgRNA is known to form multiple secondary structures, heat denaturation at 65 °C for 7 minutes is needed prior to loading into the gel to remove all secondary structures, allowing for accurate assessment of its size.
Results:

Figure 4. RNA transcription check
The gel photo above clearly indicates the successful transcription of the sgRNA and asRNA, with the size matching their expected size (sgRNA=138bp, asRNA=140bp).
Step 3 - RNA in vitro hybridization and visualization
In order to test the environment needed for in vitro hybridization, 4 different environments were tested in a combination of using 3x Hybridization Buffer (HB) and the use of initial denaturation. Regardless of the conditions being tested, the 2 RNAs are incubated together at 37°C for 45 minutes, before snap cooling to 0°C to preserve the hybridized structure and RNase degradation. (protocol we used for the experiment)
It is known that salt content is crucial to RNA duplex formation, thus, 3x HB is used to provide the necessary salt content and stabilize the RNAs. DEPC water is used as a negative control to the testing of hybridization buffer.
Since this assay is designed as an in vitro demonstration of the in vivo hybridization, the temperature of incubation is set to 37°C to simulate the in vivo environment with no initial denaturation. As a fail-safe test, the necessity for initial denaturation is also tested as the in vitro hybridization is expected to be less efficient without the Hfq Chaperone. Therefore, denaturing the RNAs before slow cooling will remove the RNAs secondary structure while promoting the complementation and hybridization during slow cooling.
A non-denaturing 2% MOPS agarose gel is then used to visualize the hybridization.
Results:
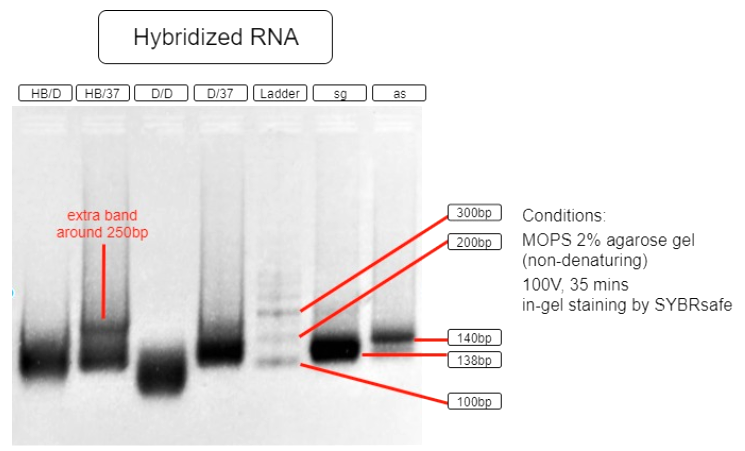
Figure 5. RNA hybridization assay
| HB/D | Hybridization Buffer + denature + slow cool* + 37°C incubation |
| HB/37 | Hybridization Buffer + slow cool* + 37°C incubation |
| D/D | DEPC H2O + denature + slow cool* +37°C incubation |
| D/37 | DEPC H2O + 37°C incubation |
| L | Ambion Century™ - Plus RNA Markers |
| sg | sgRNA BBa_K3017061 (138bp) |
| as | asRNA BBa_K3017060 (140bp) |
*slow cool is done by cooling the RNA sample using thermo cycler at 5°C per 10 sec until incubation temperature
As shown in the above gel photo (Figure 5), only lane AB/37 shows an extra band which appears to be around 250bp when compared to the ladder. This band indicates the hybridization of sgRNA and asRNA and is therefore running slower in agarose gel.
This result shows that salt content provided by the hybridization buffer (60mM KCl, 6mM HEPES, 0.2mM MgCl2 at 1x), which is similar to that of a cellular salinity, is crucial to the hybridization. This result also indicates that the RNAs are able to hybridize with each other in 37°C incubation with no initial denaturation needed. These results suggest that the RNAs are capable of hybridizing in vivo. With endogenous expression of Hfq protein in E.coli, the asRNA will also stabilize due to Spot 42 and attract other ssRNAs to hybridize.
Thus, we suspect the in vivo hybridization efficiency would be higher than our results.
References:
[1] Y. J. Lee, A. Hoynes-Oconnor, M. C. Leong, and T. S. Moon, “Programmable control of bacterial gene expression with the combined CRISPR and antisense RNA system,” Nucleic Acids Research, vol. 44, no. 5, pp. 2462–2473, Feb. 2016.
[2] C. Anders, O. Niewoehner, A. Duerst, and M. Jinek, “Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease,” Nature, vol. 513, no. 7519, pp. 569–573, 2014.
[3] S. H. Sternberg, S. Redding, M. Jinek, E. C. Greene, and J. A. Doudna, “DNA Interrogation by the CRISPR RNA-Guided Endonuclease Cas9,” Biophysical Journal, vol. 106, no. 2, 2014.
[4] T. Karvelis, G. Gasiunas, A. Miksys, R. Barrangou, P. Horvath, and V. Siksnys, “crRNA and tracrRNA guide Cas9-mediated DNA interference inStreptococcus thermophilus,” RNA Biology, vol. 10, no. 5, pp. 841–851, 2013.
[5] T. Møller, T. Franch, P. Højrup, D. R. Keene, H. P. Bächinger, R. G. Brennan, and P. Valentin-Hansen, “Hfq,” Molecular Cell, vol. 9, no. 1, pp. 23–30, 2002.
[6] G. M. Cech, A. Szalewska-Pałasz, K. Kubiak, A. Malabirade, W. Grange, V. Arluison, and G. Węgrzyn, “The Escherichia Coli Hfq Protein: An Unattended DNA-Transactions Regulator,” Frontiers in Molecular Biosciences, vol. 3, 2016.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]