Difference between revisions of "Part:BBa K2912017"
m (→Usage and Characterization) |
BingruFeng (Talk | contribs) |
||
| (22 intermediate revisions by 4 users not shown) | |||
| Line 16: | Line 16: | ||
<partinfo>BBa_K2912017 SequenceAndFeatures</partinfo> | <partinfo>BBa_K2912017 SequenceAndFeatures</partinfo> | ||
| − | + | ==SZU-China 2019 iGEM team== | |
| − | SZU-China 2019 iGEM team was going to find a suicide switch inside the E. coli that can break the whole body of the bacteria leading to the release of RNAi molecules transcribed from E. coli inducing by IPTG or some other else. Therefore, we required a useful mechanism. Fortunately, we finally found the | + | SZU-China 2019 iGEM team was going to find a suicide switch inside the E. coli that can break the whole body of the bacteria leading to the release of RNAi molecules transcribed from E. coli inducing by IPTG or some other else. Therefore, we required a useful mechanism. Fortunately, we finally found the '''Refractile inclusion bodies (R-bodies)'''to kill the E. coli, causing the inclusion to flow out of the plasma membrane so that we can get the RNAi molecules transcribed by E. coli (Fig.1). |
<div> | <div> | ||
| − | <center><html><img src='https://2019.igem.org/wiki/images/4/40/T--SZU-CHINA--exp4.jpg' style="width: | + | <center><html><img src='https://2019.igem.org/wiki/images/4/40/T--SZU-CHINA--exp4.jpg' style="width:40%;margin:0 auto"> |
| − | <center><html><img src='https://2019.igem.org/wiki/images/4/48/T--SZU-CHINA--exp5.jpg' style="width: | + | <center><html><img src='https://2019.igem.org/wiki/images/4/48/T--SZU-CHINA--exp5.jpg' style="width:40%;margin:0 auto"> |
| − | <center><html><img src='https://2019.igem.org/wiki/images/2/23/T--SZU-CHINA--exp6.jpg' style="width: | + | <center><html><img src='https://2019.igem.org/wiki/images/2/23/T--SZU-CHINA--exp6.jpg' style="width:40%;margin:0 auto"> |
<center> Fig.1 Synthesis of our RNAi molecules </center></html></center> | <center> Fig.1 Synthesis of our RNAi molecules </center></html></center> | ||
</div> | </div> | ||
====Refractile inclusion bodies==== | ====Refractile inclusion bodies==== | ||
| − | Refractile inclusion bodies, known as R bodies, are produced by only a few species of bacteria. These inclusion bodies are highly insoluble protein ribbons, typically seen coiled into cylindrical structures within the cell[1]. R-bodies are produced by Paramecium endosymbionts belonging to the genus Caedibacter. These intracellular bacteria confer upon their hosts, a phenomenon called the killer trait[2]. The R bodies of | + | Refractile inclusion bodies, known as R bodies, are produced by only a few species of bacteria. These inclusion bodies are highly insoluble protein ribbons, typically seen coiled into cylindrical structures within the cell[1]. R-bodies are produced by Paramecium endosymbionts belonging to the genus Caedibacter. These intracellular bacteria confer upon their hosts, a phenomenon called the killer trait[2]. The R bodies of ''C. taeniospiralis'' are type 51. They are about '''0.5 μm wide''', have '''a maximum length of 20 μm''', and 13 nm thick, possess acute angles at each end and '''unroll''' in a telescopic fashion when '''exposed to a pH of 6.5 or lower'''. These proteinaceous ribbons are rolling up inside the cell to form '''a hollow cylinder''' about '''0.5 μm in diameter''' and 0.5 μm long[3]. |
====Usage and Characterization==== | ====Usage and Characterization==== | ||
| Line 33: | Line 33: | ||
<div> | <div> | ||
| − | <center><html><img src=' | + | <center><html><img src='https://2019.igem.org/wiki/images/3/3e/T--SZU-CHINA--R-body-Plasmid.png' style="width:40%;margin:0 auto"> |
<center> Fig.2 Plasmid construction of R-body </center></html></center> | <center> Fig.2 Plasmid construction of R-body </center></html></center> | ||
</div> | </div> | ||
| − | The attenuator was designed to control the expression of R-body via the concentration of Tryptophan. When the concentration of Trp is under 0.3%, R-body begins to express. Hence, after our RNAi molecules are adequately transcribed inside E. coli, the R-body proteins can be induced to translate to form the R-bodies rolled-up inside the bacteria. Then, we will change the pH of the media to induce the R-bodies to unroll and crack the E. coli after the R-bodies are fully expressed. | + | The attenuator was designed to control the expression of R-body via the concentration of Tryptophan(Fig.3). When the concentration of Trp is under 0.3%, R-body begins to express. Hence, after our RNAi molecules are adequately transcribed inside E. coli, the R-body proteins can be induced to translate to form the R-bodies rolled-up inside the bacteria(Fig.4). Then, we will change the pH of the media to induce the R-bodies to unroll and crack the E. coli after the R-bodies are fully expressed. |
<div> | <div> | ||
| − | <center><html><img src=' | + | <center><html><img src='https://2019.igem.org/wiki/images/e/e3/T--SZU-CHINA--R-body-Trp.png' style="width:40%;margin:0 auto"> |
<center> Fig.3 The Regulation of Trp Attenuator </center></html></center> | <center> Fig.3 The Regulation of Trp Attenuator </center></html></center> | ||
</div> | </div> | ||
<div> | <div> | ||
| − | <center><html><img src=' | + | <center><html><img src='https://2019.igem.org/wiki/images/f/fc/T--SZU-CHINA--R-body-pH.png' style="width:40%;margin:0 auto"> |
<center> Fig.4 The Regulation of pH on R-Bodies </center></html></center> | <center> Fig.4 The Regulation of pH on R-Bodies </center></html></center> | ||
</div> | </div> | ||
| − | The R-bodies proteins were predicted to fully translate after 44 minutes. Click [https://2019.igem.org/Team:SZU-China/Model SZU-China 2019 Model_R-body] to see more. | + | The R-bodies proteins were predicted to fully translate after 44 minutes. Click [https://2019.igem.org/Team:SZU-China/Model SZU-China 2019 Model_R-body]to see more. |
SZU-China 2019 iGEM team has synthesized the R-bodies and taken the scanning electron microscope pictures of the R-bodies under pH=7 and pH=6 (Fig.5,6). | SZU-China 2019 iGEM team has synthesized the R-bodies and taken the scanning electron microscope pictures of the R-bodies under pH=7 and pH=6 (Fig.5,6). | ||
| Line 54: | Line 54: | ||
<div> | <div> | ||
<center> | <center> | ||
| − | <html><img src='https://2019.igem.org/wiki/images/d/d5/T--SZU-CHINA--R-body1.png' style="width: | + | <html><img src='https://2019.igem.org/wiki/images/d/d5/T--SZU-CHINA--R-body1.png' style="width:40%;margin:0 auto"> |
| − | + | <center> Fig.5 SEM pictures of the R-bodies under pH=7 </center></html></center> | |
| − | <center> Fig.5 | + | |
</div> | </div> | ||
| + | |||
| + | <div> | ||
| + | <center> | ||
| + | <html><img src='https://2019.igem.org/wiki/images/b/b3/T--SZU-CHINA--R-body-3.png' style="width:40%;margin:0 auto"> | ||
| + | <html><img src='https://2019.igem.org/wiki/images/e/ee/T--SZU-CHINA--R-body-4.png' style="width:40%;margin:0 auto"> | ||
| + | <center> Fig.6 SEM pictures of the R-bodies under pH=6 </center></html></center> | ||
| + | </div> | ||
| + | |||
| + | ==SZU-China 2022 iGEM team== | ||
| + | We carefully examine the properties of R-body (Refractile inclusion bodies) and find some interesting points. The natural R-body gene cluster has four genes in the order of RebA-RebB-RebD-RebC. Reb A can act as a scaffolding protein to facilitate the major polymerization process; Reb B is the major structural subunit of the R body; a polypeptide as small as RebC, by binding to RebA or RebB, induces a conformational change that enables the modifying protein to modify RebAB; and RebD is not transcribed or translated in E. coli. | ||
| + | |||
| + | <center>[[File:K4286504-figure1-A.png|400px]]</center> | ||
| + | |||
| + | <center>[[File:K4286504-figure1-B.png|400px]]</center> | ||
| + | |||
| + | <center><b>Figure 1. R-body gene cluster before and after modification</b></center> | ||
| + | |||
| + | First of all, RebD is not transcribed or translated in E. coli, of course, it may be due the frequency of expression is very low or only expressed for a short time, it is not recognized by the test. Experiments have shown that R-body can be synthesized in E. coli in the apparent absence of RebD,indicating that RebD may not be essential for R-body production. Therefore, we propose the first improvement, which is to delete RebD to ensure the simplification of the gene cluster of R-body; Secondly, the main structural component of our R-body protein complex is RebB, and RebB is in the second place in the natural gene cluster. We plan to move RebB to the first place, so as to improve the expression of RebB, and make the synthesis and assembly of R-body in E. coli more efficient. | ||
| + | |||
| + | By regulating pH, R-body extension can physically disrupt the entire body of the bacterium, thereby releasing shRNA molecules transcribed by E. coli. In summary, we obtained a modified version of the R-body gene cluster, in which the gene are arranged in the order of RebB-RebA-RebC. | ||
| + | |||
| + | ====Assembly==== | ||
| + | |||
| + | We constructed the recombinant vectors [RebABDC]-pRSFDuet1 and [RebBAC]-pRSFDuet1, through which we compared the R-body production between the part of 2019 SZU-China and our improved part. | ||
| + | |||
| + | <center>[[File:K4286504-figure2.png|600px]]</center> | ||
| + | |||
| + | <center><b>Figure 2. Plasmid [RebABDC]-pRSFDuet1 (with original version of R-body gene cluster)</b></center> | ||
| + | |||
| + | <center>[[File:K4286504-figure3.png|600px]]</center> | ||
| + | |||
| + | <center><b>Figure 3. Plasmid [RebBAC]-pRSFDuet1 (with modified version of R-body gene cluster)</b></center> | ||
| + | |||
| + | We transferred the recombinant vector into E.coli Ht115(DE3). Transformants were clearly visible on the culture medium after 16 hours of incubation at 37℃. | ||
| + | |||
| + | <center>[[File:K4286504-figure4.png|600px]]</center> | ||
| + | |||
| + | <center><b>Figure 4. Colonial morphology of E.coli transformants (left: [RebABDC]-pRSFDuet1 transformants; right: [RebBAC]-pRSFDuet1 transformants).</b></center> | ||
| + | |||
| + | We selected 8 single colonies on each culture medium and carried out colony PCR for plasmid amplification. The theoretical length of the amplified product was 1730bp (RebABDC) and 1340bp (RebBAC). Electrophoresis was performed in a 1% agarose gel. The results showed that all the colonies were positive transformants, which indicated that the recombinant vector was successfully transformed. | ||
| + | |||
| + | <center>[[File:K4286504-figure5.png|600px]]</center> | ||
| + | |||
| + | <center><b>Figure 5. 1% agarose gel stained with RebABDC and RebBAC integration of pRSFDuet1 in E.coli Ht115(DE3) was checked by PCR. The capitalized word with a number represents the sample we choose. M: 2000bp Marker. RA1~8: RebABDC transformants. RB1~8: RebBAC transformants.</b></center> | ||
| + | |||
| + | After enlarged production of recombinant plasmids, we conducted double restriction enzyme digestion for further verification. Restriction enzyme NdeI & HindIII was used for digestion. For [RebABDC]-pRSFDuet1, the theoretical sizes of bands are 449bp and 4094bp; for [RebBAC]-pRSFDuet1, the theoretical sizes of bands are 98bp and 4196bp. Electrophoresis was performed in a 1% agarose gel. The results showed successful double-enzyme digestion and correct plasmid extraction. | ||
| + | |||
| + | <center>[[File:K4286504-figure6.png|600px]]</center> | ||
| + | |||
| + | <center><b>Figure 6. 1% agarose gel stained with RebABDC and RebBAC integration of pRSFDuet1 in E.coli Ht115(DE3) was checked by restriction enzyme digestion.</b></center> | ||
| + | |||
| + | <center>M: 10000bp Marker. pRA1, pRA2, pRB1, pRB2: the plasmids without enzyme digestion, which showed closed circular plasmid DNA (cc DNA) and open circular plasmid DNA (oc DNA). s-pRA1, s-pRA2, s-pRB1, s-pRB2: plasmid digested by restriction enzyme HindIII, which showed linear plasmid DNA (Linear DNA) with theoretical band size 4543bp and 4294bp for RA and RB, respectively. d-pRA1, d-pRA2, d-pRB1, d-pRB2: plasmid digested by restriction enzyme NdeI and HindIII, which showed theoretical band size 449bp and 4094bp for RA, 98bp and 4196bp for RB.</center> | ||
| + | |||
| + | ====Characterization==== | ||
| + | |||
| + | We compared our improved R-body part with the former R-body part designed by 2019 SZU-iGEM. Since the structure of R-body is complicated, it is difficult to purify them by protein labels for verification. Therefore, we characterized the effect and number of Escherichia coli treated with R-body lysis to reflect the R-body protein yield. | ||
| + | |||
| + | We treated transformant solution with 2% L-arabinose solution for 4 hours to induce R-body expression. Then we used 1 mol/L acetic acid solution to induce R-body from coiling to unwrapping, within which Escherichia coli can be lysed. We set the following induction conditions, and measured OD 600 of E.coli solution before and after induction under each condition. | ||
| + | |||
| + | <center><b>Table 1. Induction conditions of E.coli transformants</b></center> | ||
| + | |||
| + | <center>[[File:K4286504-table1.jpg|600px]]</center> | ||
| + | |||
| + | We compared Escherichia coli in LB liquid media that had induced R-body production with those that had not. The following bar graph shows the decrease in OD 600 of liquid media before and after acid treatment (Fig. 7). After L-arabinose treatment, R-body were produced in E.coli, which lysed a large number of E.coli after acid induction. In groups with L-arabinose treatment, OD 600 changed significantly, especially in groups RB1++ and RB2++. When E.coli were not treated with L-arabinose, R-body was absent in vivo, within which acid treatment had almost no effect on OD 600. In groups without L-arabinose treatment, only a small amount of E.coli died, which was due to acidic environment. It can be indicated that we successfully induced R-body production and E.coli lysis, using L-arabinose and acetic acid, respectively. | ||
| + | |||
| + | <center>[[File:K4286504-figure7.jpg|600px]]</center> | ||
| + | |||
| + | <center><b>Figure 7. OD 600 of E.coli In LB Liquid Media Before And After Treatments.</b></center> | ||
| + | |||
| + | <center>RA1, RA2: E.coli transformants containing [RebABDC]-pRSFDuet1 plasmids. RB1, RB2: E.coli transformants containing [RebBAC]-pRSFDuet1 plasmids. ++: E.coli treated with L-arabinose and acetic acid. -+: E.coli treated with acetic acid only.</center> | ||
| + | |||
===References=== | ===References=== | ||
Latest revision as of 14:46, 11 October 2022
Refractile inclusion bodies_ Caedibacter
Refractile inclusion bodies SZU-China 2019 iGEM team was going to find a suicide switch inside the E coli that can break the whole body of the bacteria leading to the release of RNAi molecules transcribed from E coli inducing by IPTG or some other else. Therefore, we were in need the useful mechanism. Fortunately, we finally found the Refractile inclusion bodies (R-bodies) to kill the E coli, causing the inclusion to flow out of the plasma membrane, so that we can get the RNAi molecules transcribed by E coli. Refractile inclusion bodies, known as R bodies, are produced by only a few species of bacteria. These inclusion bodies are highly insoluble protein ribbons, typically seen coiled into cylindrical structures within the cell[1]. R-bodies are produced by Paramecium endosymbionts belonging to the genus Caedibacter. These intracellular bacteria confer upon their hosts a phenomenon called the killer trait[2]. This is one of the DNA sequences for the R body locus (reb) from Caedibacter taeniospiralis. The R bodies of C. taeniospiralis are type 51. They are about 0.5 μm wide, have a maximum length of 20 μm, and 13 nm thick, possess acute angles at each end, and unroll in a telescopic fashion when exposed to a pH of 6.5 or lower. These proteinaceous ribbons are rolling up inside the cell to form a hollow cylinder about 0.5 μm in diameter and 0.5 μm long. Sequence and Features
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 15
Illegal NheI site found at 38 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
SZU-China 2019 iGEM team
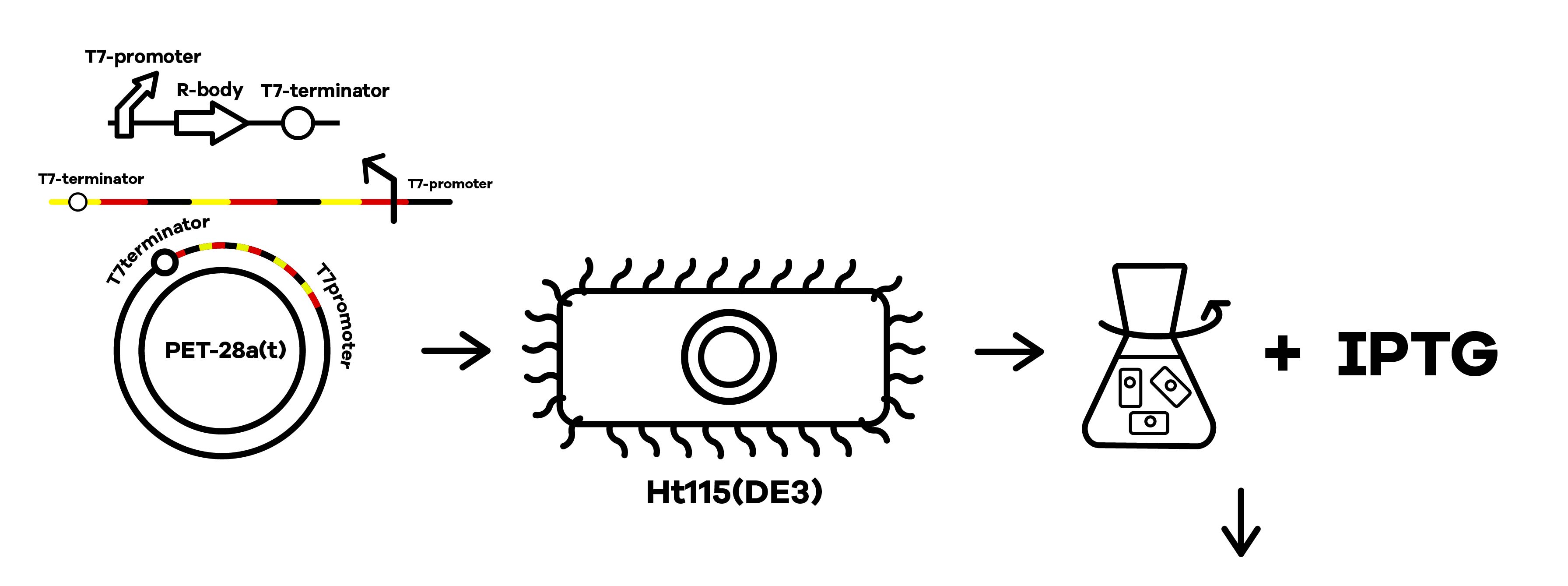
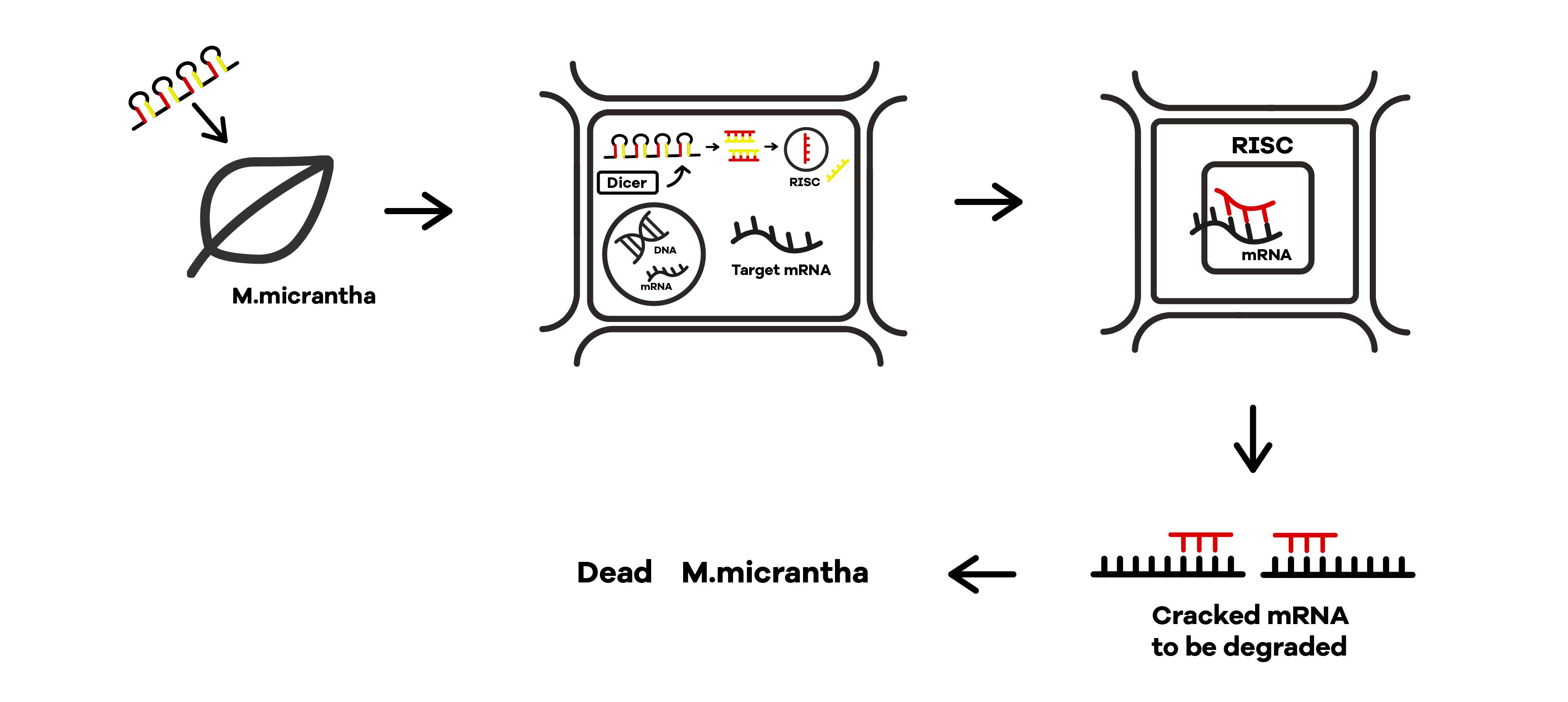
SZU-China 2019 iGEM team was going to find a suicide switch inside the E. coli that can break the whole body of the bacteria leading to the release of RNAi molecules transcribed from E. coli inducing by IPTG or some other else. Therefore, we required a useful mechanism. Fortunately, we finally found the Refractile inclusion bodies (R-bodies)to kill the E. coli, causing the inclusion to flow out of the plasma membrane so that we can get the RNAi molecules transcribed by E. coli (Fig.1).
Refractile inclusion bodies
Refractile inclusion bodies, known as R bodies, are produced by only a few species of bacteria. These inclusion bodies are highly insoluble protein ribbons, typically seen coiled into cylindrical structures within the cell[1]. R-bodies are produced by Paramecium endosymbionts belonging to the genus Caedibacter. These intracellular bacteria confer upon their hosts, a phenomenon called the killer trait[2]. The R bodies of C. taeniospiralis are type 51. They are about 0.5 μm wide, have a maximum length of 20 μm, and 13 nm thick, possess acute angles at each end and unroll in a telescopic fashion when exposed to a pH of 6.5 or lower. These proteinaceous ribbons are rolling up inside the cell to form a hollow cylinder about 0.5 μm in diameter and 0.5 μm long[3].
Usage and Characterization
SZU-China 2019 iGEM team constructed a plasmid with BBa_K1475900 (Promoter),BBa_K2912014 (Attenuator),BBa_K2912000 (Reb A),BBa_K2912001 (Reb B),BBa_K2912003 (Reb D),BBa_K2912002 (Reb C),BBa_B0010 (Terminator)(Fig.2).

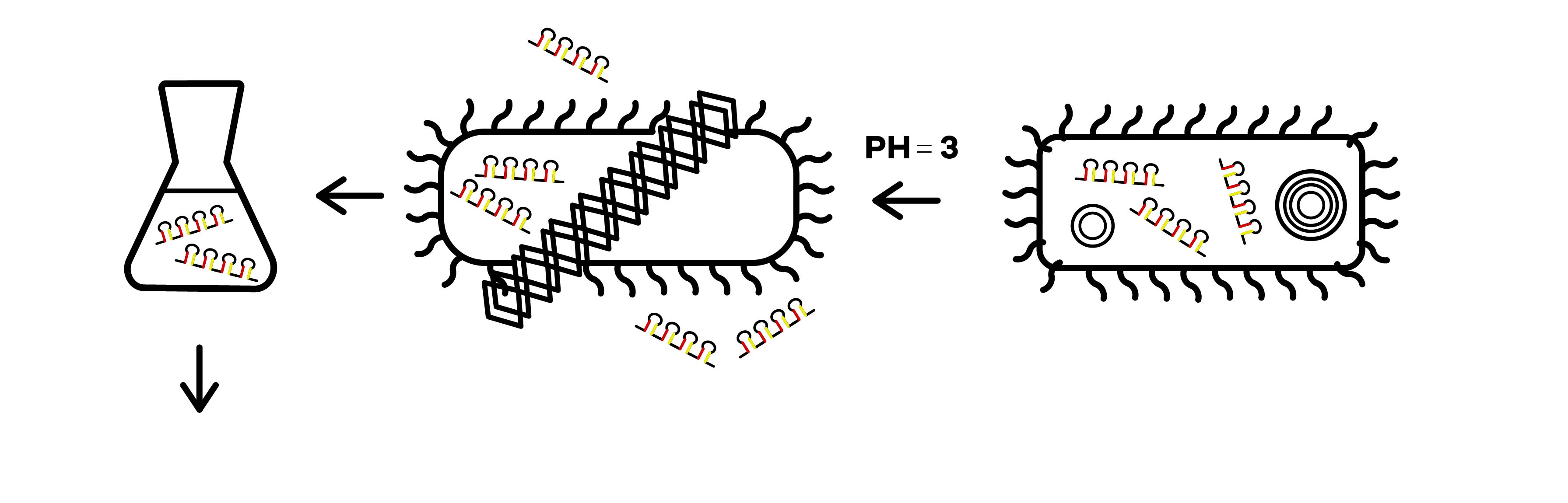
The attenuator was designed to control the expression of R-body via the concentration of Tryptophan(Fig.3). When the concentration of Trp is under 0.3%, R-body begins to express. Hence, after our RNAi molecules are adequately transcribed inside E. coli, the R-body proteins can be induced to translate to form the R-bodies rolled-up inside the bacteria(Fig.4). Then, we will change the pH of the media to induce the R-bodies to unroll and crack the E. coli after the R-bodies are fully expressed.


The R-bodies proteins were predicted to fully translate after 44 minutes. Click SZU-China 2019 Model_R-bodyto see more.
SZU-China 2019 iGEM team has synthesized the R-bodies and taken the scanning electron microscope pictures of the R-bodies under pH=7 and pH=6 (Fig.5,6).



SZU-China 2022 iGEM team
We carefully examine the properties of R-body (Refractile inclusion bodies) and find some interesting points. The natural R-body gene cluster has four genes in the order of RebA-RebB-RebD-RebC. Reb A can act as a scaffolding protein to facilitate the major polymerization process; Reb B is the major structural subunit of the R body; a polypeptide as small as RebC, by binding to RebA or RebB, induces a conformational change that enables the modifying protein to modify RebAB; and RebD is not transcribed or translated in E. coli.


First of all, RebD is not transcribed or translated in E. coli, of course, it may be due the frequency of expression is very low or only expressed for a short time, it is not recognized by the test. Experiments have shown that R-body can be synthesized in E. coli in the apparent absence of RebD,indicating that RebD may not be essential for R-body production. Therefore, we propose the first improvement, which is to delete RebD to ensure the simplification of the gene cluster of R-body; Secondly, the main structural component of our R-body protein complex is RebB, and RebB is in the second place in the natural gene cluster. We plan to move RebB to the first place, so as to improve the expression of RebB, and make the synthesis and assembly of R-body in E. coli more efficient.
By regulating pH, R-body extension can physically disrupt the entire body of the bacterium, thereby releasing shRNA molecules transcribed by E. coli. In summary, we obtained a modified version of the R-body gene cluster, in which the gene are arranged in the order of RebB-RebA-RebC.
Assembly
We constructed the recombinant vectors [RebABDC]-pRSFDuet1 and [RebBAC]-pRSFDuet1, through which we compared the R-body production between the part of 2019 SZU-China and our improved part.


We transferred the recombinant vector into E.coli Ht115(DE3). Transformants were clearly visible on the culture medium after 16 hours of incubation at 37℃.

We selected 8 single colonies on each culture medium and carried out colony PCR for plasmid amplification. The theoretical length of the amplified product was 1730bp (RebABDC) and 1340bp (RebBAC). Electrophoresis was performed in a 1% agarose gel. The results showed that all the colonies were positive transformants, which indicated that the recombinant vector was successfully transformed.

After enlarged production of recombinant plasmids, we conducted double restriction enzyme digestion for further verification. Restriction enzyme NdeI & HindIII was used for digestion. For [RebABDC]-pRSFDuet1, the theoretical sizes of bands are 449bp and 4094bp; for [RebBAC]-pRSFDuet1, the theoretical sizes of bands are 98bp and 4196bp. Electrophoresis was performed in a 1% agarose gel. The results showed successful double-enzyme digestion and correct plasmid extraction.

Characterization
We compared our improved R-body part with the former R-body part designed by 2019 SZU-iGEM. Since the structure of R-body is complicated, it is difficult to purify them by protein labels for verification. Therefore, we characterized the effect and number of Escherichia coli treated with R-body lysis to reflect the R-body protein yield.
We treated transformant solution with 2% L-arabinose solution for 4 hours to induce R-body expression. Then we used 1 mol/L acetic acid solution to induce R-body from coiling to unwrapping, within which Escherichia coli can be lysed. We set the following induction conditions, and measured OD 600 of E.coli solution before and after induction under each condition.

We compared Escherichia coli in LB liquid media that had induced R-body production with those that had not. The following bar graph shows the decrease in OD 600 of liquid media before and after acid treatment (Fig. 7). After L-arabinose treatment, R-body were produced in E.coli, which lysed a large number of E.coli after acid induction. In groups with L-arabinose treatment, OD 600 changed significantly, especially in groups RB1++ and RB2++. When E.coli were not treated with L-arabinose, R-body was absent in vivo, within which acid treatment had almost no effect on OD 600. In groups without L-arabinose treatment, only a small amount of E.coli died, which was due to acidic environment. It can be indicated that we successfully induced R-body production and E.coli lysis, using L-arabinose and acetic acid, respectively.

References
[1] Pond FR, Gibson I, Lalucat J, et al. R-body-producing Bacteria.[J]. Microbiol Rev, 1989, 53(1): 25-67.
[2] Matsuoka JI, Ishizuna F, Kurumisawa K, et al. Stringent Expression Control of Pathogenic R-body Production in Legume Symbiontazorhizobium Caulinodans[J]. Mbio, 2017, 8(4): 0-17.
[3] Heruth DP, Pond FR, Dilts JA, et al. Characterization of Genetic Determinants for R Body Synthesis and Assembly in Caedibacter Taeniospiralis 47 and 116.[J]. Journal of Bacteriology, 1994, 176(12): 3559-3567.
