Difference between revisions of "Part:BBa K3239007"
BrianSebZhou (Talk | contribs) |
Nicole Guo (Talk | contribs) (→Usage and Biology) |
||
| (18 intermediate revisions by 2 users not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K3239007 short</partinfo> | <partinfo>BBa_K3239007 short</partinfo> | ||
| − | yEFGP3 reporter gene expressed by | + | ''yEFGP3'' reporter gene expressed by ''P<sub>AOX1''. |
===Usage and Biology=== | ===Usage and Biology=== | ||
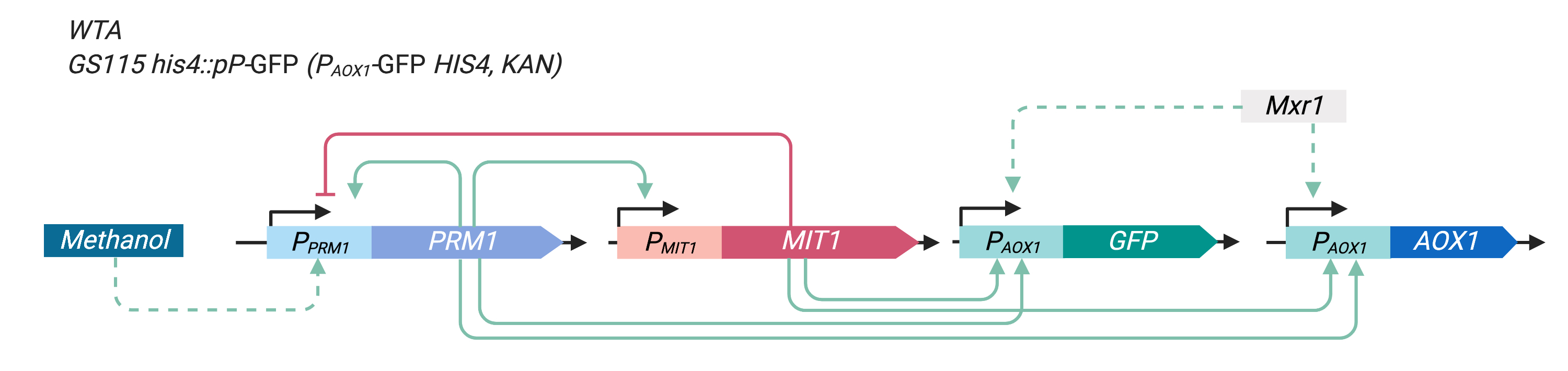
| − | This construct is the control used in our experiment. After homologous recombination into wild type P. pastoris, the EGFP expressed shall serve as an indicator of AOX1 expression levels. Note that the original | + | This construct is the control used in our experiment. After homologous recombination into wild type ''P. pastoris'', the EGFP protein expressed shall serve as an indicator of ''AOX1'' expression levels. Note that the original ''P<sub>AOX1''-''AOX1'' copy remains intact after the recombination. |
| + | |||
| + | [[File: pAOX1-GFP.png|400px|thumb|Figure 1. Illustration of the ''P<sub>AOX1''-GFP construct]] | ||
| + | |||
| + | |||
| + | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Latest revision as of 06:22, 18 October 2019
Status: 500
Content-type: text/html
Software error:
Global symbol "$groups" requires explicit package name at /websites/parts.igem.org/cgi/lib/User.pm line 84. Global symbol "$groups" requires explicit package name at /websites/parts.igem.org/cgi/lib/User.pm line 84. Global symbol "$groups" requires explicit package name at /websites/parts.igem.org/cgi/lib/User.pm line 85. Compilation failed in require at /websites/parts.igem.org/cgi/lib/IconBar.pm line 12. BEGIN failed--compilation aborted at /websites/parts.igem.org/cgi/lib/IconBar.pm line 12. Compilation failed in require at /websites/parts.igem.org/cgi/lib/BBWeb.pm line 9. BEGIN failed--compilation aborted at /websites/parts.igem.org/cgi/lib/BBWeb.pm line 9. Compilation failed in require at /websites/parts.igem.org/cgi/lib/Change.pm line 8. BEGIN failed--compilation aborted at /websites/parts.igem.org/cgi/lib/Change.pm line 8. Compilation failed in require at /websites/parts.igem.org/cgi/lib/Part.pm line 12. BEGIN failed--compilation aborted at /websites/parts.igem.org/cgi/lib/Part.pm line 12. Compilation failed in require at /websites/parts.igem.org/cgi/partsdb/putout.cgi line 8. BEGIN failed--compilation aborted at /websites/parts.igem.org/cgi/partsdb/putout.cgi line 8.
For help, please send mail to this site's webmaster, giving this error message and the time and date of the error.
yEFGP3 reporter gene expressed by PAOX1.
Usage and Biology
This construct is the control used in our experiment. After homologous recombination into wild type P. pastoris, the EGFP protein expressed shall serve as an indicator of AOX1 expression levels. Note that the original PAOX1-AOX1 copy remains intact after the recombination.
Sequence and Features
Status: 500
Content-type: text/html
Software error:
Global symbol "$groups" requires explicit package name at /websites/parts.igem.org/cgi/lib/User.pm line 84. Global symbol "$groups" requires explicit package name at /websites/parts.igem.org/cgi/lib/User.pm line 84. Global symbol "$groups" requires explicit package name at /websites/parts.igem.org/cgi/lib/User.pm line 85. Compilation failed in require at /websites/parts.igem.org/cgi/lib/IconBar.pm line 12. BEGIN failed--compilation aborted at /websites/parts.igem.org/cgi/lib/IconBar.pm line 12. Compilation failed in require at /websites/parts.igem.org/cgi/lib/BBWeb.pm line 9. BEGIN failed--compilation aborted at /websites/parts.igem.org/cgi/lib/BBWeb.pm line 9. Compilation failed in require at /websites/parts.igem.org/cgi/lib/Change.pm line 8. BEGIN failed--compilation aborted at /websites/parts.igem.org/cgi/lib/Change.pm line 8. Compilation failed in require at /websites/parts.igem.org/cgi/lib/Part.pm line 12. BEGIN failed--compilation aborted at /websites/parts.igem.org/cgi/lib/Part.pm line 12. Compilation failed in require at /websites/parts.igem.org/cgi/partsdb/putout.cgi line 8. BEGIN failed--compilation aborted at /websites/parts.igem.org/cgi/partsdb/putout.cgi line 8.
For help, please send mail to this site's webmaster, giving this error message and the time and date of the error.