Difference between revisions of "Part:BBa K3021004"
| (17 intermediate revisions by the same user not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K3021004 short</partinfo> | <partinfo>BBa_K3021004 short</partinfo> | ||
| − | This is a new composite part designed to allow Laccase secretion from our engineered E.coli strain through embedded the new secretion peptide, novel signal peptide 4 (NSP4). Laccase is an important enzyme, which would degrade different chemicals. The Laccase sequence in this part is based on the laccase1326 cDNA sequence from previous study, which was shown to have great tolerant to different harsh conditions commonly found in polluted water. Secretion of heterologous proteins into Escherichia coli cell culture medium offers significant advantages for downstream processing over production. NSP4 is utilizing part of the Sec-dependent (Sec) secretion pathway, the general secretion E. coli route. | + | This is a new composite part designed to allow Laccase secretion from our engineered E.coli strain through embedded the new secretion peptide, novel signal peptide 4 (NSP4). Laccase is an important enzyme, which would degrade different chemicals. The Laccase sequence in this part is based on the laccase1326 cDNA sequence from previous study, which was shown to have great tolerant to different harsh conditions commonly found in polluted water. Secretion of heterologous proteins into Escherichia coli cell culture medium offers significant advantages for downstream processing over production. NSP4 is utilizing part of the Sec-dependent (Sec) secretion pathway, the general secretion E. coli route. |
| − | + | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| Line 17: | Line 16: | ||
<partinfo>BBa_K3021004 parameters</partinfo> | <partinfo>BBa_K3021004 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | |||
| + | |||
| + | <html> | ||
| + | <style> | ||
| + | .legend { | ||
| + | font-size: .9em; | ||
| + | } | ||
| + | p { | ||
| + | text-align: justify; | ||
| + | } | ||
| + | .content img { | ||
| + | max-height:80vh; | ||
| + | max-width:100%; | ||
| + | margin: 0 auto; | ||
| + | clear: both; | ||
| + | } | ||
| + | </style> | ||
| + | <div class="content"> | ||
| + | <h2>Successful expression of K3021004</h2> | ||
| + | |||
| + | <p>In order to validate the protein expression of K3021004 (with Lac1326 laccase), K863000 and K3021002 (with K863030 laccase), we performed SDS-PAGE (coomassie blue staining) and Western blot analysis. To make Lac1326, K863000 and K3021002(with K863030 laccase) easily detectable via Western Blot, we designed our constructs to include a “His tag” which is targeted by a commercially available antibody. With the SDS-PAGE and western blot (anti-his for his-tagged protein), we found successful expression of K3021004 (with Lac1326 laccase), K863000 and K3021002 (with K863030 laccase).</p> | ||
| + | |||
| + | <p>Before performing the SDS-PAGE, we grew up our engineered E. coli cells containing a plasmid with K3021004 (with Lac1326 laccase), K863000 and K3021002 (K863030) under control of the T7 promoter. Cells were induced with 0.4mM, 1.0mM or 1.2mM IPTG until either OD0.4 or OD1.2 (wavelength: 600nm) at 25°C or 30°C. After expression, cells were then lysed according to our protein extraction protocol, and supernatant and pellet were collected for the SDS-PAGE.</p> | ||
| + | For more experimental details of the Western Blot and SDS-PAGE, please see <a href="https://2019.igem.org/Team:PuiChing_Macau/Protocol">protocol</a>. | ||
| + | |||
| + | <img src="https://2019.igem.org/wiki/images/6/64/T--PuiChing_Macau--results_5.png"> | ||
| + | <div class="legend">Figure 1. Western Blot results of inducible expression of K3021004 (Lac1326), K3021002 (K863030) and K863000. | ||
| + | <br>(a) | ||
| + | <ul>Lane1: pET-11a empty vector; | ||
| + | <li>Lane2: pET-11a empty vector+IPTG induction</li> | ||
| + | <li>Lane3: K3021004 (Lac1326) colony1;</li> | ||
| + | <li>Lane4:K3021004 (Lac1326) colony1+IPTG induction</li> | ||
| + | <li>Lane5: K3021004 (Lac1326) colony2;</li> | ||
| + | <li>Lane6:K3021004 (Lac1326) colony2+IPTG induction</li> | ||
| + | <li>Lane7: K3021004 (Lac1326) colony3;</li> | ||
| + | <li>Lane8:K3021004 (Lac1326) colony3+IPTG induction</li> | ||
| + | <li>Lane9: K3021002 (K863030) colony1;</li> | ||
| + | <li>Lane10: K3021002 (K863030) colony1+IPTG induction</li> | ||
| + | </ul> | ||
| + | (b) | ||
| + | <ul> | ||
| + | <li>Lane1: K863000 colony2;</li> | ||
| + | <li>Lane2: K863000 colony2+IPTG induction</li> | ||
| + | <li>Lane3: K863000 colony3;</li> | ||
| + | <li>Lane4: K863000 colony3+IPTG induction</li> | ||
| + | <li>Lane5: K3021002 (K863030) colony1;</li> | ||
| + | <li>Lane6: K3021002 (K863030) colony1+IPTG induction</li> | ||
| + | <li>Lane7: K3021002 (K863030) colony2;</li> | ||
| + | <li>Lane8: K3021002 (K863030) colony2+IPTG induction</li> | ||
| + | <li>Lane9: K3021002 (K863030) colony3;</li> | ||
| + | <li>Lane10: K3021002 (K863030) colony3+IPTG induction</li> | ||
| + | </ul></div> | ||
| + | <p>As shown in Figure 1, Wetern Blot was performed to validate the presence of the his-tagged K3021004 (with Lac1326 laccase), K863000 and K3021002 (with K863030 laccase) protein. Anti-RNA polymersase beta was used as a loading control. | ||
| + | At 58 kD, we found a band in lane 3 to lane 8 which corresponds to the approximate expected size of K3021004 (with Lac1326 laccase). We also found a band at 64kD in lanes 9 and 10, which is the expected size of K3021002 (with K863030 laccase).</p> | ||
| + | |||
| + | <h2>IPTG Induction of K3021004</h2> | ||
| + | |||
| + | <img src="https://2019.igem.org/wiki/images/8/8f/T--PuiChing_Macau--results_6.png"> | ||
| + | <div class="legend">Figure 2. The figure represents the expression of different laccases under a variable of IPTG. The protein expression is calculated by the ratio of anti-His and anti-RNA polymerase beta (Loading control).</div> | ||
| + | <p>As we used IPTG inducible promoter (T7) in our study, we further analyze the results of Western Blot to compare the laccase expression with/without IPTG. As shown in Figure 6, the expressions of different laccases under a variable of IPTG were compared. K3021004 (Lac1326), K3021002 (K863030) and K863000 with 1mM IPTG have exhibited higher expression levels (33.97, 41.66 and 28.75 folds, respectively) than K3021004 (Lac1326), K3021002 (K863030) and K863000 without IPTG (5.53, 21.99 and 15.53 folds). There was enhanced expression of the laccases with IPTG as expected.</p> | ||
| + | |||
| + | |||
| + | <h2>Expression of K3021004 under different conditions</h2> | ||
| + | <img src="https://2019.igem.org/wiki/images/1/12/T--PuiChing_Macau--results_7.png"> | ||
| + | <img src="https://2019.igem.org/wiki/images/2/20/T--PuiChing_Macau--results_8.png"> | ||
| + | <div class="legend">Figure 3. The figure represents expression of laccases under differnt conditions. Anti-RNA polymerase beta was used as loading control. | ||
| + | <ul> | ||
| + | <li>Lane1: K3021004 (Lac1326): OD=0.4, IPTG=0.4mM, 30°C</li> | ||
| + | <li>Lane2: K3021004 (Lac1326): OD=0.4, IPTG=0.4mM, 25 °C</li> | ||
| + | <li>Lane3:K3021004 (Lac1326): OD=0.4, IPTG=1mM, 30 °C</li> | ||
| + | <li>Lane4:K3021004 (Lac1326): OD=1.2, IPTG=1mM, 30 °C</li> | ||
| + | <li>Lane5:K863000: OD=0.4, IPTG=0.4mM, 30 °C</li> | ||
| + | <li>Lane6:K863000: OD=0.4, IPTG=0.4mM, 25 °C</li> | ||
| + | <li>Lane7:K863000: OD=0.4, IPTG=1mM, 30 °C</li> | ||
| + | <li>Lane8:K863000: OD=1.2, IPTG=1mM, 30 °C</li> | ||
| + | <li>Lane9:K3021002 (K863030): OD=0.4, IPTG=0.4mM, 30 °C</li> | ||
| + | <li>Lane10:K3021002 (K863030): OD=0.4, IPTG=0.4mM, 25 °C</li> | ||
| + | <li>Lane11:K3021002 (K863030): OD=0.4, IPTG=1mM, 30 °C</li> | ||
| + | <li>Lane12:K3021002 (K863030): OD=1.2, IPTG=1mM, 30 °C</li> | ||
| + | <li>Lane13: pET-11a empty vector without IPTG</li> | ||
| + | </ul> | ||
| + | <p>After confirming the expression of laccases and confirming the laccases under T7 promoter can be regulated by IPTG, we further tested how laccases vary across different conditions (cell growth phase, temperatures, IPTG concentration), as shown in Figure 3.</p> | ||
| + | |||
| + | |||
| + | <img src="https://2019.igem.org/wiki/images/6/62/T--PuiChing_Macau--results_19.png"> | ||
| + | <div class="legend">Figure 4. The figure represents the expression of K3021004 (Lac1326) under different temperatures, E. coli concentration and iPTG concentration. The protein expression is calculated by the ratio of anti-his and anti-RNA polymerase beta (Loading control).</div> | ||
| + | <p>The figure compares the expression of K3021004 (Lac1326)under different conditions including temperature, E coli and IPTG concentration. K3021004 (Lac1326) expressed under OD=0.4, IPTG=0.4mM at 25°C, has displayed the highest fold changes of 24.58 compared with the pET-11a empty vector, signifying that OD=0.4, IPTG=0.4mM and 25°C is the best expression temperature for K3021004 (Lac1326).</p> | ||
| + | |||
| + | <h2>Functional Secretion Signal Peptides</h2> | ||
| + | <img src="https://2019.igem.org/wiki/images/a/a2/T--PuiChing_Macau--results_11.png"> | ||
| + | <div class="legend">Figure 5. Expression of laccase with his-tag and secretion signal peptide in cell-free medium | ||
| + | <ul> | ||
| + | <li>Lane1: K3021004 (Lac1326): OD=0.4, IPTG=0.4mM, 30°C</li> | ||
| + | <li>Lane2: K3021004 (Lac1326): OD=0.4, IPTG=0.4mM, 25°C</li> | ||
| + | <li>Lane3:K3021004 (Lac1326): OD=0.4, IPTG=1mM, 30°C</li> | ||
| + | <li>Lane4:K3021004 (Lac1326): OD=1.2, IPTG=1mM, 30°C</li> | ||
| + | <li>Lane5:K3021002 (K863030): OD=0.4, IPTG=0.4mM, 30°C</li> | ||
| + | <li>Lane6:K3021002 (K863030): OD=0.4, IPTG=0.4mM, 25°C</li> | ||
| + | <li>Lane7:K3021002 (K863030): OD=0.4, IPTG=1mM, 30°C</li> | ||
| + | <li>Lane8:K3021002 (K863030): OD=1.2, IPTG=1mM, 30°C</li> | ||
| + | <li>Lane9: pET-11a empty vector without IPTG</li> | ||
| + | </ul></div> | ||
| + | |||
| + | <img src="https://2019.igem.org/wiki/images/4/47/T--PuiChing_Macau--results_12.png"> | ||
| + | <div class="legend">Figure 6. The figure shows the laccases (K3021004 (Lac1326) and K3021002 (K863030)) secretion from the engineered E. coli by Nickel-pulldown assay. Tested with one-sample student t-test. **=p value=0.002739 | ||
| + | Since we designed a secret form for the two laccases, we hypothesized that there would be his-tagged protein in the medium in which two genetically engineered E. coli were cultured. | ||
| + | First of all, we obtained the cell-free medium by spinning down the cells. We then used the Nickel-pulldown Assay and Western Blot to confirm the expression of his-tagged laccase. The results (Figure 10 and Figure 11) showed that the fold changes of K3021004 (with Lac1326 laccase) and K3021002 (with K863030 laccase) was 389.5 and 62.12 (compared to pET-11a control), indicating expression of K3021004 (with Lac1326 laccase) (significant) and K3021002 (with K863030 laccase) in the medium . | ||
| + | |||
| + | |||
| + | <h2>Dye Decolorization</h2> | ||
| + | <img src="https://2019.igem.org/wiki/images/8/8c/T--PuiChing_Macau--results_13.jpeg"> | ||
| + | <img src="https://2019.igem.org/wiki/images/9/92/T--PuiChing_Macau--results_14.jpeg"> | ||
| + | <div class="legend">Figure 7. Representative experiment of dye decolorization</div> | ||
| + | <img src="https://2019.igem.org/wiki/images/f/f4/T--PuiChing_Macau--results_15.png"> | ||
| + | <div class"legend">Figure 8. Decolorization of Indigo Carmine (initial concentration of 10 mg/L) by different laccases (OD 0.4, IPTG 0.4mM, 25 °C) at different times with ABTS.</div> | ||
| + | <p>In order to investigate the biodegradable reactivity of the laccases, dye (Indigo Carmine) decolorization experiments were conducted (Figure 7).</p> | ||
| + | <p>As shown in Figure 8, the laccase K863000 from Bielefied-Germany 2012 was the most efficient enzyme with 81.09% removal of Indigo Carmine (initial concentration: 10 mg/L) after 96 hours incubation with ABTS at 37°C comparing to K3021004 (Lac1326) and K3021002 (K863030) which decolorized 47.62% and 22.01% of Indigo Carmine after 96 hours.</p> | ||
| + | |||
| + | <h2>Successful EDC degradation</h2> | ||
| + | <img src="https://2019.igem.org/wiki/images/2/27/T--PuiChing_Macau--results_16.png"> | ||
| + | <div class="legend">Figure 9. Chromatogram of Beta-estradiol (Quantitative ion pair: 297.049/106.906) in various protein (a) Blank in ACN (b) K3021002 (K863030) (c) K863000 (d) K3021004 (Lac1326) (e) pET-11a</div> | ||
| + | |||
| + | <img src="https://2019.igem.org/wiki/images/a/ab/T--PuiChing_Macau--results_17.png"> | ||
| + | |||
| + | <div class="legend">Figure 10. Liquid Chromatography–Mass Spectrometry analysis for Beta-estradiol degradation by different laccases expressed under the condition of OD=0.4, IPTG=0.4mM and 25°C.</div> | ||
| + | |||
| + | <p>In order to validate our concept of using laccases to degrade Endocrine Disrupting Chemicals, biodegradation test were conducted. We used beta-estradiol (one of the EDCs) as our the target substrate. After incubation of laccases with beta-estradiol, we analyzed the amount beta-estradiol with Liquid Chromatography-Mass Spectrometry.</p> | ||
| + | <p>As shown in Figure 9 and Figure 10, the concentration of beta-estradiol decreased after treatment with K3021004 (wiht Lac1326 laccase), K863000 and K3021002 (with K863030 laccase) proteins, compared with pET-11a (empty vector), which successfully demonstrated our concept of utilizing K3021004 (with Lac1326 laccase) for EDC degradation. Among all of the laccases, K863000 and K3021004 (with Lac1326 laccase) had respectively displayed the lowest and second lowest intensity of 2137.657 and 2421.598, signifying K863000 possesses the highest biodegradation activity of all.</p> | ||
| + | <p>Moreover, we found that the results of dye decolorization and EDC degradation are consistent. Protein K863000 has the highest laccase activity, followed by K3021004 (wiht Lac1326 laccase) (OD0.4, IPTG=0.4mM, 25℃;) and then K3021002 (with K863030 laccase). </p> | ||
| + | |||
| + | |||
| + | |||
| + | </div> | ||
| + | </html> | ||
Latest revision as of 00:36, 22 October 2019
NSP4-Laccase1326-HisTag
This is a new composite part designed to allow Laccase secretion from our engineered E.coli strain through embedded the new secretion peptide, novel signal peptide 4 (NSP4). Laccase is an important enzyme, which would degrade different chemicals. The Laccase sequence in this part is based on the laccase1326 cDNA sequence from previous study, which was shown to have great tolerant to different harsh conditions commonly found in polluted water. Secretion of heterologous proteins into Escherichia coli cell culture medium offers significant advantages for downstream processing over production. NSP4 is utilizing part of the Sec-dependent (Sec) secretion pathway, the general secretion E. coli route. Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1493
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 1007
Illegal BamHI site found at 1526
Illegal XhoI site found at 227 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 136
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 862
Successful expression of K3021004
In order to validate the protein expression of K3021004 (with Lac1326 laccase), K863000 and K3021002 (with K863030 laccase), we performed SDS-PAGE (coomassie blue staining) and Western blot analysis. To make Lac1326, K863000 and K3021002(with K863030 laccase) easily detectable via Western Blot, we designed our constructs to include a “His tag” which is targeted by a commercially available antibody. With the SDS-PAGE and western blot (anti-his for his-tagged protein), we found successful expression of K3021004 (with Lac1326 laccase), K863000 and K3021002 (with K863030 laccase).
Before performing the SDS-PAGE, we grew up our engineered E. coli cells containing a plasmid with K3021004 (with Lac1326 laccase), K863000 and K3021002 (K863030) under control of the T7 promoter. Cells were induced with 0.4mM, 1.0mM or 1.2mM IPTG until either OD0.4 or OD1.2 (wavelength: 600nm) at 25°C or 30°C. After expression, cells were then lysed according to our protein extraction protocol, and supernatant and pellet were collected for the SDS-PAGE.
For more experimental details of the Western Blot and SDS-PAGE, please see protocol.
(a)
- Lane1: pET-11a empty vector;
- Lane2: pET-11a empty vector+IPTG induction
- Lane3: K3021004 (Lac1326) colony1;
- Lane4:K3021004 (Lac1326) colony1+IPTG induction
- Lane5: K3021004 (Lac1326) colony2;
- Lane6:K3021004 (Lac1326) colony2+IPTG induction
- Lane7: K3021004 (Lac1326) colony3;
- Lane8:K3021004 (Lac1326) colony3+IPTG induction
- Lane9: K3021002 (K863030) colony1;
- Lane10: K3021002 (K863030) colony1+IPTG induction
- Lane1: K863000 colony2;
- Lane2: K863000 colony2+IPTG induction
- Lane3: K863000 colony3;
- Lane4: K863000 colony3+IPTG induction
- Lane5: K3021002 (K863030) colony1;
- Lane6: K3021002 (K863030) colony1+IPTG induction
- Lane7: K3021002 (K863030) colony2;
- Lane8: K3021002 (K863030) colony2+IPTG induction
- Lane9: K3021002 (K863030) colony3;
- Lane10: K3021002 (K863030) colony3+IPTG induction
As shown in Figure 1, Wetern Blot was performed to validate the presence of the his-tagged K3021004 (with Lac1326 laccase), K863000 and K3021002 (with K863030 laccase) protein. Anti-RNA polymersase beta was used as a loading control. At 58 kD, we found a band in lane 3 to lane 8 which corresponds to the approximate expected size of K3021004 (with Lac1326 laccase). We also found a band at 64kD in lanes 9 and 10, which is the expected size of K3021002 (with K863030 laccase).
IPTG Induction of K3021004

As we used IPTG inducible promoter (T7) in our study, we further analyze the results of Western Blot to compare the laccase expression with/without IPTG. As shown in Figure 6, the expressions of different laccases under a variable of IPTG were compared. K3021004 (Lac1326), K3021002 (K863030) and K863000 with 1mM IPTG have exhibited higher expression levels (33.97, 41.66 and 28.75 folds, respectively) than K3021004 (Lac1326), K3021002 (K863030) and K863000 without IPTG (5.53, 21.99 and 15.53 folds). There was enhanced expression of the laccases with IPTG as expected.
Expression of K3021004 under different conditions


- Lane1: K3021004 (Lac1326): OD=0.4, IPTG=0.4mM, 30°C
- Lane2: K3021004 (Lac1326): OD=0.4, IPTG=0.4mM, 25 °C
- Lane3:K3021004 (Lac1326): OD=0.4, IPTG=1mM, 30 °C
- Lane4:K3021004 (Lac1326): OD=1.2, IPTG=1mM, 30 °C
- Lane5:K863000: OD=0.4, IPTG=0.4mM, 30 °C
- Lane6:K863000: OD=0.4, IPTG=0.4mM, 25 °C
- Lane7:K863000: OD=0.4, IPTG=1mM, 30 °C
- Lane8:K863000: OD=1.2, IPTG=1mM, 30 °C
- Lane9:K3021002 (K863030): OD=0.4, IPTG=0.4mM, 30 °C
- Lane10:K3021002 (K863030): OD=0.4, IPTG=0.4mM, 25 °C
- Lane11:K3021002 (K863030): OD=0.4, IPTG=1mM, 30 °C
- Lane12:K3021002 (K863030): OD=1.2, IPTG=1mM, 30 °C
- Lane13: pET-11a empty vector without IPTG
After confirming the expression of laccases and confirming the laccases under T7 promoter can be regulated by IPTG, we further tested how laccases vary across different conditions (cell growth phase, temperatures, IPTG concentration), as shown in Figure 3.

The figure compares the expression of K3021004 (Lac1326)under different conditions including temperature, E coli and IPTG concentration. K3021004 (Lac1326) expressed under OD=0.4, IPTG=0.4mM at 25°C, has displayed the highest fold changes of 24.58 compared with the pET-11a empty vector, signifying that OD=0.4, IPTG=0.4mM and 25°C is the best expression temperature for K3021004 (Lac1326).
Functional Secretion Signal Peptides

- Lane1: K3021004 (Lac1326): OD=0.4, IPTG=0.4mM, 30°C
- Lane2: K3021004 (Lac1326): OD=0.4, IPTG=0.4mM, 25°C
- Lane3:K3021004 (Lac1326): OD=0.4, IPTG=1mM, 30°C
- Lane4:K3021004 (Lac1326): OD=1.2, IPTG=1mM, 30°C
- Lane5:K3021002 (K863030): OD=0.4, IPTG=0.4mM, 30°C
- Lane6:K3021002 (K863030): OD=0.4, IPTG=0.4mM, 25°C
- Lane7:K3021002 (K863030): OD=0.4, IPTG=1mM, 30°C
- Lane8:K3021002 (K863030): OD=1.2, IPTG=1mM, 30°C
- Lane9: pET-11a empty vector without IPTG

Dye Decolorization
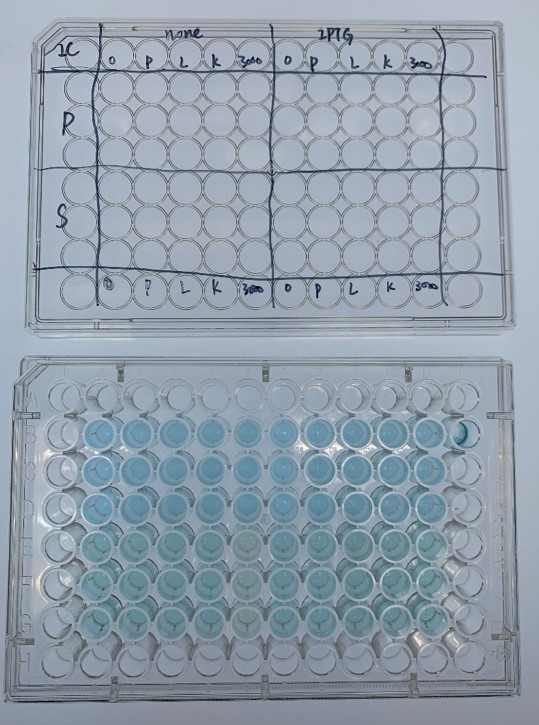
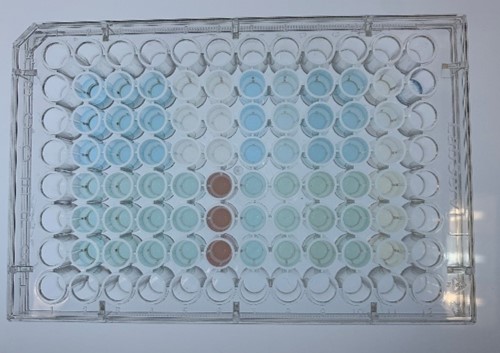
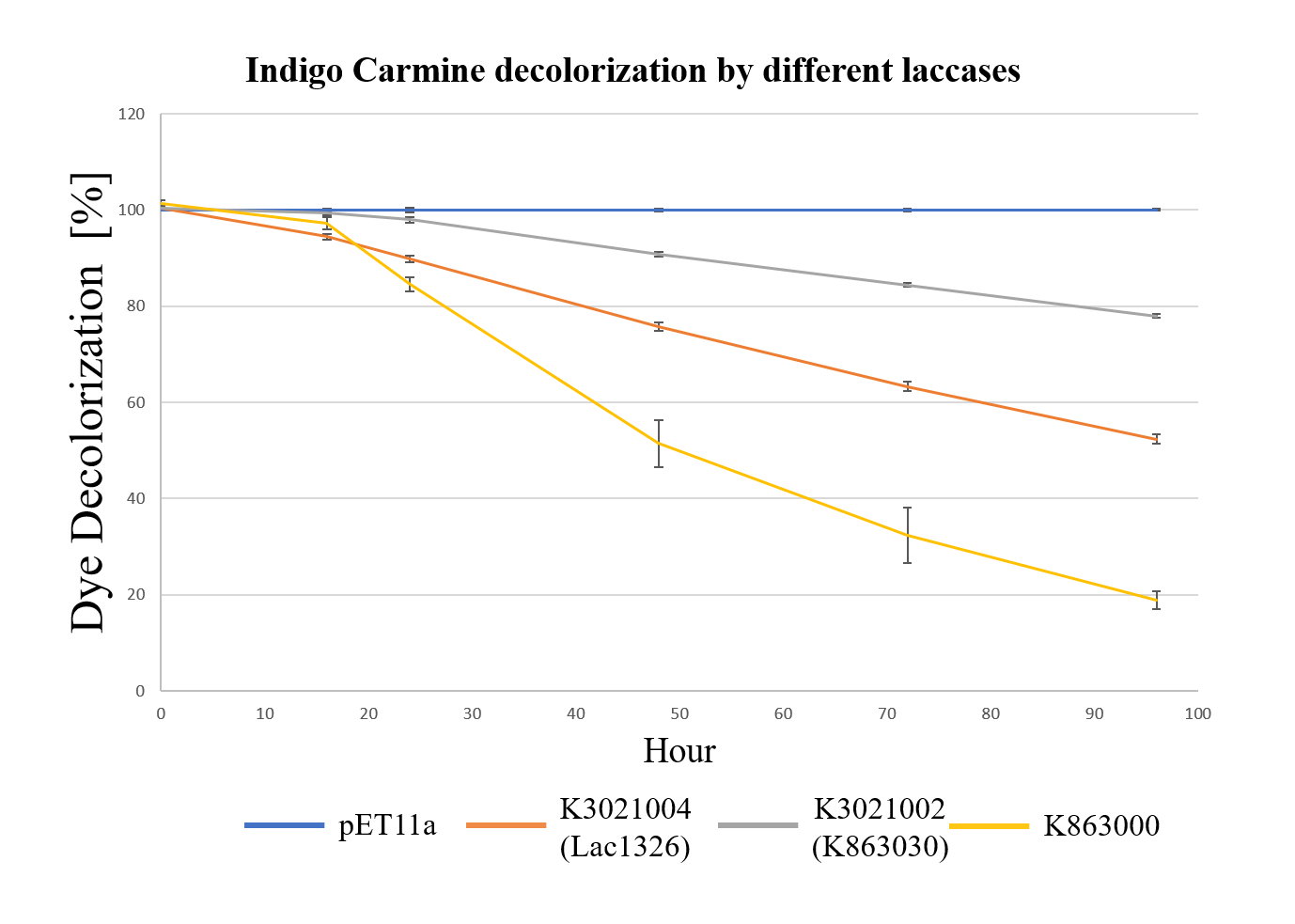
In order to investigate the biodegradable reactivity of the laccases, dye (Indigo Carmine) decolorization experiments were conducted (Figure 7).
As shown in Figure 8, the laccase K863000 from Bielefied-Germany 2012 was the most efficient enzyme with 81.09% removal of Indigo Carmine (initial concentration: 10 mg/L) after 96 hours incubation with ABTS at 37°C comparing to K3021004 (Lac1326) and K3021002 (K863030) which decolorized 47.62% and 22.01% of Indigo Carmine after 96 hours.
Successful EDC degradation
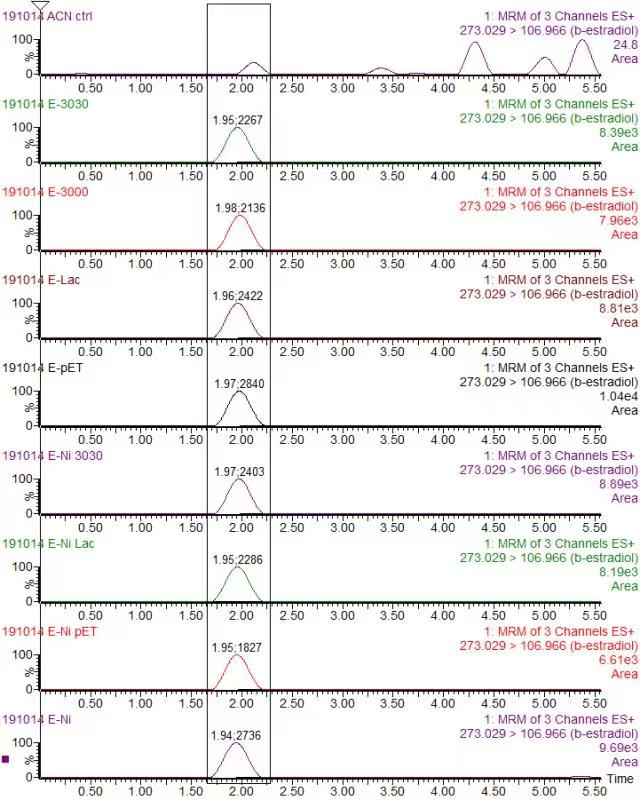
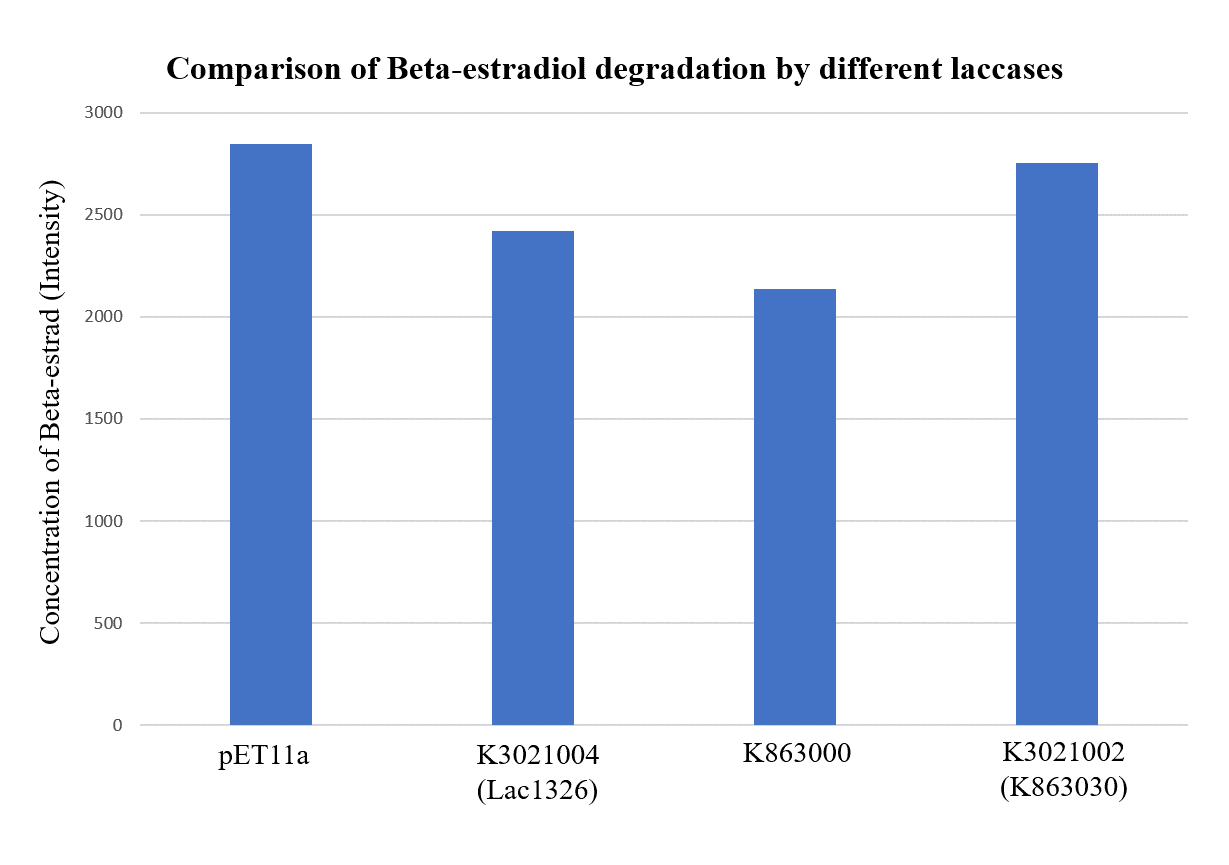
In order to validate our concept of using laccases to degrade Endocrine Disrupting Chemicals, biodegradation test were conducted. We used beta-estradiol (one of the EDCs) as our the target substrate. After incubation of laccases with beta-estradiol, we analyzed the amount beta-estradiol with Liquid Chromatography-Mass Spectrometry.
As shown in Figure 9 and Figure 10, the concentration of beta-estradiol decreased after treatment with K3021004 (wiht Lac1326 laccase), K863000 and K3021002 (with K863030 laccase) proteins, compared with pET-11a (empty vector), which successfully demonstrated our concept of utilizing K3021004 (with Lac1326 laccase) for EDC degradation. Among all of the laccases, K863000 and K3021004 (with Lac1326 laccase) had respectively displayed the lowest and second lowest intensity of 2137.657 and 2421.598, signifying K863000 possesses the highest biodegradation activity of all.
Moreover, we found that the results of dye decolorization and EDC degradation are consistent. Protein K863000 has the highest laccase activity, followed by K3021004 (wiht Lac1326 laccase) (OD0.4, IPTG=0.4mM, 25℃;) and then K3021002 (with K863030 laccase).
