Difference between revisions of "Part:BBa K143002"
m |
m |
||
| (6 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | ===<big>3' Integration Sequence for the | + | ===<big>3' Integration Sequence for the amyE Locus of ''B. subtilis''</big>=== |
| − | [[Image: | + | [[Image:IntegrationAmyE.PNG|center|800px]] |
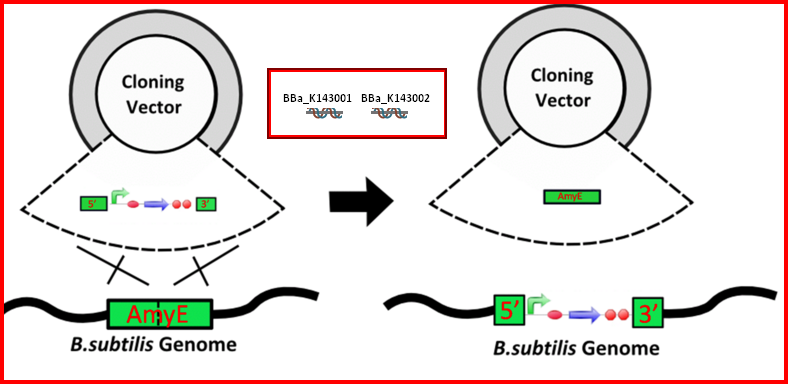
| − | Integration sequences allow DNA to be incorporated into the chromosome of a host cell at a specific locus using leading (5') and trailing (3') DNA sequences that are the same as those at a specific locus of the chromosome. The 5' integration sequence can be added to the front of a | + | Integration sequences allow DNA to be incorporated into the chromosome of a host cell at a specific locus using leading (5') and trailing (3') DNA sequences that are the same as those at a specific locus of the chromosome. The 5' integration sequence can be added to the front of a BioBrick construct and the 3' integration sequence specific for this locus (<bbpart>BBa_K143001</bbpart>) to the rear of the BioBrick construct to allow integration of the BioBrick constructs into the chromosome of the Gram-positive bacterium ''B. subtilis'' at the amyE locus. |
| + | |||
| + | The amyE locus was the first locus used for integration into ''B. subtilis'' by Shimotsu and Henner[1] and is still commonly used in vectors such as pDR111[2], pDL[3] and their derivatives. Integration at the amyE locus removes the ability of ''B. subtilis'' to break down starch, which can be assayed with iodine as described by Cutting and Vander-horn[4]. For the iGEM 2008 competition, the Imperial Team used the 5' and 3' integration sequences for the amyE locus to integrate genetic construct into the ''B. subtilis'' chromosome. | ||
| + | |||
| + | |||
| + | <!-- --> | ||
| + | <span class='h3bb'><big>'''Sequence and Features'''</big></span> | ||
| + | |||
| + | |||
| + | <partinfo>BBa_K143002 SequenceAndFeatures</partinfo> | ||
| − | |||
===References=== | ===References=== | ||
| Line 15: | Line 23: | ||
#4 Cutting, S M.; Vander-Horn, P B. Genetic analysis. In: Harwood C R, Cutting S M. , editors. Molecular biological methods for Bacillus. Chichester, England: John Wiley & Sons, Ltd.; 1990. pp. 27–74. | #4 Cutting, S M.; Vander-Horn, P B. Genetic analysis. In: Harwood C R, Cutting S M. , editors. Molecular biological methods for Bacillus. Chichester, England: John Wiley & Sons, Ltd.; 1990. pp. 27–74. | ||
</biblio> | </biblio> | ||
| + | |||
| + | |||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Latest revision as of 01:42, 30 October 2008
3' Integration Sequence for the amyE Locus of B. subtilis
Integration sequences allow DNA to be incorporated into the chromosome of a host cell at a specific locus using leading (5') and trailing (3') DNA sequences that are the same as those at a specific locus of the chromosome. The 5' integration sequence can be added to the front of a BioBrick construct and the 3' integration sequence specific for this locus (BBa_K143001) to the rear of the BioBrick construct to allow integration of the BioBrick constructs into the chromosome of the Gram-positive bacterium B. subtilis at the amyE locus.
The amyE locus was the first locus used for integration into B. subtilis by Shimotsu and Henner[1] and is still commonly used in vectors such as pDR111[2], pDL[3] and their derivatives. Integration at the amyE locus removes the ability of B. subtilis to break down starch, which can be assayed with iodine as described by Cutting and Vander-horn[4]. For the iGEM 2008 competition, the Imperial Team used the 5' and 3' integration sequences for the amyE locus to integrate genetic construct into the B. subtilis chromosome.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
References
<biblio>
- 1 pmid=3019840
- 2 pmid=14597697
- 3 Bacillus Genetic Stock Center [www.bgsc.org]
- 4 Cutting, S M.; Vander-Horn, P B. Genetic analysis. In: Harwood C R, Cutting S M. , editors. Molecular biological methods for Bacillus. Chichester, England: John Wiley & Sons, Ltd.; 1990. pp. 27–74.
</biblio>