Difference between revisions of "Part:BBa K3111011"
m (→Experimental Results) |
|||
| (27 intermediate revisions by 3 users not shown) | |||
| Line 17: | Line 17: | ||
<partinfo>BBa_K3111011 parameters</partinfo> | <partinfo>BBa_K3111011 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | |||
| + | <html> | ||
| + | </p> | ||
| + | </html> | ||
| + | __TOC__ | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| − | + | Designed Ankyrin Repeat Proteins (DARPins) are small (14-21 kDa) single-domain binding molecules which can bind to specific targets and subsequently trigger various molecular mechanisms (e.g. enzyme inhibition or protein anchoring) (2,3). They have been genetically engineered by Andreas Plückthun in 2003, as a means to manipulate the properties of natural ankyrin repeat proteins for therapeutic applications (4,5). New DARPins can be created via mutagenesis and phase display technologies, which enables the selection of highly stable, soluble, easily-expressed and efficient versions. This indicates that these binding molecules have the potential to circumvent the limitations of monoclonal antibodies and empower new therapeutic approaches (4). In short, they possess the following qualities making them ideal for therapeutic approaches: | |
| + | |||
| + | * Due to their high specificity, stability (thermodynamic), and affinity, as well as their flexible architecture, DARPins have a rigid body-binding mode. They are stable in human blood serum and can be engineered so as not to contain T-cell epitopes (5). | ||
| + | * Their unique pharmacokinetic properties can be altered upon fusion with half-life extending molecules (5,7). | ||
| + | * They can be produced rapidly and cost-effectively (i.e. in E. coli) (as opposed to antibodies that are expensive, have a tedious manufacturing process and need to be humanized before administration) (8). | ||
| + | * Multi-specific DARPin constructs can be made by genetic fusion while retaining similar binding properties as single-domain DARPins, meaning that they can provide greater clinical benefit by overcoming the limitations of conventional therapeutic approaches, that typically target a single disease pathway (many diseases result from the dysregulation of multiple pathways). DARPin technology can be harnessed to rapidly generate many different “multi-DARPins” which enable targeting of multiple disease pathways (1,6,7). | ||
| + | * DARPins have been shown to be very stable and show less tendencies for aggregation, unlike single chain variable fragments since they do not possess cysteines. | ||
| − | DARPin929 has been one of the most frequently used to evaluate probes for molecular imaging. It particularly targets human epidermal growth factor 2 (HER2) whose overexpression is associated with breast cancer and gastroesophageal cancer. | + | DARPin929, in particular, has been one of the most frequently used to evaluate probes for molecular imaging. It particularly targets human epidermal growth factor 2 (HER2) whose overexpression is associated with breast cancer and gastroesophageal cancer (9). It has been shown to bind to its target with nanomolar affinities (10,11). |
===Functionalisation=== | ===Functionalisation=== | ||
| Line 33: | Line 44: | ||
In order to assess the feasibility of fusing DARPin929 to either encapsulin, we created protein models using PyMol (GUI and manual edits), Discovery Studio (structure quality), GROMACS (energy minimisation of created dimers) and atomium (rebuilding the full encapsulin capsules). | In order to assess the feasibility of fusing DARPin929 to either encapsulin, we created protein models using PyMol (GUI and manual edits), Discovery Studio (structure quality), GROMACS (energy minimisation of created dimers) and atomium (rebuilding the full encapsulin capsules). | ||
| − | + | Figures 1 and 2 show the full capsule models of <i>T. maritima</i> (Fig. 1) and <i>M. xanthus</i> (Fig. 2) encapsulins with DARPins bound to outer terminal of each monomer. | |
| + | |||
| + | [[File:T--UCL--T maritima encapsulin-darpins.png|700px|thumb|center|Figure 1. A preview of the full <i>T. maritima</i> encapsulin with DARPins clearly resolved. The DARPins from two di-pentamers are highlighted.]] | ||
| + | |||
| + | [[File:T--UCL--Mx encapsulin-darpins.png|700px|thumb|center|Figure 2. Part of <i>M. xanthus</i> encapsulin surface. Represented are the interactions between DARPins linked to the three equivalent encapsulin chains (A, B and P, respectively).]] | ||
| + | |||
| + | The clashes present in Figure 2 are an artefact of building the model and would not be seen <i>in vivo</i>. However, the overall space for the DARPins to resolve without clashes and rotate to bind HER2 is evidently bigger with the <i>T. maritima</i> encapsulin. | ||
As a result we conclude that fusing DARPin929 to the <i>T. maritima</i> encapsulin is more readily feasible. We demonstrate this fusion in BBa_K3111501. | As a result we conclude that fusing DARPin929 to the <i>T. maritima</i> encapsulin is more readily feasible. We demonstrate this fusion in BBa_K3111501. | ||
| Line 45: | Line 62: | ||
====Confirmation of assembly==== | ====Confirmation of assembly==== | ||
| − | Using composite part <partinfo>BBa_K3111501</partinfo> we investigated the ability of DARPin929 to be fused onto T. maritima encapsulin without hindering its assembly. We first demonstrated that this fusion protein would express and purify without cleavage by running it on an SDS gel (Figure | + | Using composite part <partinfo>BBa_K3111501</partinfo> we investigated the ability of DARPin929 to be fused onto <i>T. maritima</i> encapsulin without hindering its assembly. We first demonstrated that this fusion protein would express and purify without cleavage by running it on an SDS gel (Figure 4). From this we could observe that the encapsulin became strongly insoluble after the addition of DARPin929, however, a small fraction of it could still be purified, as seen in lane 2 and 3 on the gel (~51 kDa). We did not observe any smaller bands (besides host cell protein at ~15 kDa), indicating that the DARPin was not cleaved off. |
| − | [[Image:TmEncH_DARPin2.png|500px|thumb|center|'''Figure | + | [[Image:TmEncH_DARPin2.png|500px|thumb|center|'''Figure 4:''' SDS PAGE of <i>T. maritima</i> encapsulin monomer_DARPin929_StrepII. Purified protein is highlighted in the by the red box. M: PageRuler<sup>TM</sup> Prestained Protein Ladder S: Cell lysate Soluble Fragment, I: Insoluble cell lysate fragment, L: Load, W: Wash, 1-3: Elution 1-3.]] |
| − | To observe assembly under non reducing environment, we concentrated the purified sample from elution 2 and used it for Transmission Electron Microscopy imaging and non-reducing PAGE gel. | + | To observe assembly under non-reducing environment, we concentrated the purified sample from elution 2 and used it for Transmission Electron Microscopy imaging and non-reducing PAGE gel. |
| − | [[Image:TmEncH_DARPin3.png|600px|thumb|center|'''Figure | + | [[Image:TmEncH_DARPin3.png|600px|thumb|center|'''Figure 5:''' a) Transmission Electron Microscopy of assembled <i>T. maritima</i> encapsulin_DARPin929_StrepII; Scalebar: 50 nm, b) Native PAGE gel of assembled <i>T. maritima</i> encapsulin_DARPin929_StrepII.]] |
| − | Figure | + | Figure 5 (a) shows the TEM image obtained. We could clearly observe assembled encapsulins thus concluding that although expression of the protein was mostly insoluble, the soluble fragment contained fully assembled encapsulins. The native PAGE gel observed in Figure 5 (b) let us confirm that the DARPin did not get cleaved-off during assembly, as we observed a significant increase in band size from the TmEnc lane to the TmEnc_DARPin929. |
After these observations we decided that assembly was not very critically affected and moved to cargo loading in part <partinfo>BBa_K3111502</partinfo>. | After these observations we decided that assembly was not very critically affected and moved to cargo loading in part <partinfo>BBa_K3111502</partinfo>. | ||
| Line 61: | Line 78: | ||
Furthermore, we investigated the impact of fusing DARPin929 on cargo loading into the encapsulin by evaluating two different DARPin display strategies. First, <partinfo>BBa_K3111502</partinfo> showed the loading of mini singlet oxyged generator (miniSOG) into an encapsulin which has a DARPin on every monomer, whereas, <partinfo>BBa_K3111503</partinfo> had a mixture of encapsulin monomer with and without a DARPin. | Furthermore, we investigated the impact of fusing DARPin929 on cargo loading into the encapsulin by evaluating two different DARPin display strategies. First, <partinfo>BBa_K3111502</partinfo> showed the loading of mini singlet oxyged generator (miniSOG) into an encapsulin which has a DARPin on every monomer, whereas, <partinfo>BBa_K3111503</partinfo> had a mixture of encapsulin monomer with and without a DARPin. | ||
| − | [[Image:TmEncH_loading.png|400px|thumb|center|'''Figure | + | [[Image:TmEncH_loading.png|400px|thumb|center|'''Figure 6:''' Quantification of <i>T. maritima</i> encapsulin loading. Error bars show 95% confidence interval.]] |
| − | Figure | + | Figure 6 shows the cargo loading capacity obtained from producing the multicomponent drug delivery platform using the aforementioned cloning strategies. Loading capacity for <partinfo>BBa_K3111502</partinfo> was estimated to be 13.61+-0.36 molecules of miniSOG. In contrast, <partinfo>BBa_K3111503</partinfo> was shown to be loading 8.19±0.21 miniSOG molecules per encapsulin. We hypothesise that this difference is caused by the surface display of DARPins slowing down the assembly of encapsulins and giving the targeting peptide present on miniSOG more time to form hydrophobic interactions to the inner shell of encapsulin monomers. Analysis on the methodology used to calculate the number of loaded cargo proteins can be found on <partinfo>BBa_K3111402</partinfo> registry page. |
===Evaluation of binding=== | ===Evaluation of binding=== | ||
| Line 69: | Line 86: | ||
To successfully display a DARPin on the encapsulin surface, it was required to fuse it downstream of the encapsulin sequence. However, we could not find any recorded cases of fusing a DARPin on the C-terminus of another protein. We hyphotesised that C-terminal fusion could obstruct the binding site of the DARPin, therefore, we decided to test this with composite parts <partinfo>BBa_K3111201</partinfo> and <partinfo>BBa_K3111202</partinfo>. These parts featured DARPin929 fused to the red fluorescent protein mScarlet via 6-aa flexible linker in two different orientations. We tested the constructs on HER2 overexpressing SK-BR-3 breast adenocarcinoma cells, to which DARPin929 can bind to with nanomolar affinities. | To successfully display a DARPin on the encapsulin surface, it was required to fuse it downstream of the encapsulin sequence. However, we could not find any recorded cases of fusing a DARPin on the C-terminus of another protein. We hyphotesised that C-terminal fusion could obstruct the binding site of the DARPin, therefore, we decided to test this with composite parts <partinfo>BBa_K3111201</partinfo> and <partinfo>BBa_K3111202</partinfo>. These parts featured DARPin929 fused to the red fluorescent protein mScarlet via 6-aa flexible linker in two different orientations. We tested the constructs on HER2 overexpressing SK-BR-3 breast adenocarcinoma cells, to which DARPin929 can bind to with nanomolar affinities. | ||
| − | Our preliminary experiments (found on the composite part pages: <partinfo>BBa_K3111201</partinfo> and <partinfo>BBa_K3111202</partinfo>) determined that the optimal conditions for the binding of DARPin929 were 37 °C and 3 μM with no significant difference observed between 2% or 20% oxygen. In order to validate the binding specificity of DARPins, we conducted further control mammalian cell culture experiments by applying the optimal conditions. These experiments were done to firstly make sure that proteins without fused DARPins would not get uptaken by the cells or bind to their surface. This was done by applying rTurboGFP at the same concentration. Secondly to observe the specificity of DARPin and subsequently our drug delivery platform we then applied the DARPin + mScarlet fusions, on Mesenchymal stem cells (MSCs) which do not express HER2 receptors. Additionally, we wanted to observe the interaction between T. maritima encapsulins and SK-BR-3 cells for future experiments. Thus we manufactured encapsulins without DARPins (<partinfo>BBa_K3111104</partinfo>) but surface displayed iLOV, a cyan fluorescent protein and applied it at the same concentration as the previous constructs. | + | Our preliminary experiments (found on the composite part pages: <partinfo>BBa_K3111201</partinfo> and <partinfo>BBa_K3111202</partinfo>) determined that the optimal conditions for the binding of DARPin929 were 37 °C and 3 μM with no significant difference observed between 2% or 20% oxygen. In order to validate the binding specificity of DARPins, we conducted further control mammalian cell culture experiments by applying the optimal conditions. These experiments were done to firstly make sure that proteins without fused DARPins would not get uptaken by the cells or bind to their surface. This was done by applying rTurboGFP at the same concentration. Secondly to observe the specificity of DARPin and subsequently our drug delivery platform we then applied the DARPin + mScarlet fusions, on Mesenchymal stem cells (MSCs) which do not express HER2 receptors. Additionally, we wanted to observe the interaction between <i>T. maritima</i> encapsulins and SK-BR-3 cells for future experiments. Thus we manufactured encapsulins without DARPins (<partinfo>BBa_K3111104</partinfo>) but surface displayed iLOV, a cyan fluorescent protein and applied it at the same concentration as the previous constructs. |
| − | [[Image:DARPin3.png|800px|thumb|center|'''Figure | + | [[Image:DARPin3.png|800px|thumb|center|'''Figure 7:''' Confocal microscopy of controls post incubation of 6-well plate at 37 °C and 20% oxygen for an hour; Scalebar: 200 μm. mScarlet_DARPin929 denotes <partinfo>BBa_K3111201</partinfo>, DARPin929_mScarlet denotes <partinfo>BBa_K3111202</partinfo> and <i>T.maritima</i> + iLOV denotes <partinfo>BBa_K3111104</partinfo>.]] |
| − | From Figure | + | From Figure 7, we observed no fluorescence thus no binding or uptake of both rTurboGFP and <i>Thermotoga maritima</i> encapsulin fused with iLOV. Thus, we confirm that in the absence of DARPins fusion proteins cannot bind or get internalised. Moreover, we observe no red fluorescence on the MSCs indicating the specificity of DARPin only for HER2 receptors, thus confirming the potential for targeted delivery platforms by using DARPin. |
| − | To obtain greater accuracy of binding percentage we proceeded with flow cytometry. Thus, we dislodged the cells from their growth wells using EDTA, while not affecting the binding of the DARPin and the HER2 receptor, and then used the sample for flow cytometry. We used the flow cytometer (BD Accuri C6 Plus) configured to observe the fluorescence within the mScarlet and CFP regions. | + | To obtain greater accuracy of binding percentage we proceeded with flow cytometry. Thus, we dislodged the cells from their growth wells using EDTA, while not affecting the binding of the DARPin and the HER2 receptor, and then used the sample for flow cytometry. We used the flow cytometer (BD Accuri<sup>TM</sup> C6 Plus) configured to observe the fluorescence within the mScarlet and CFP regions. |
| − | [[Image:DARPin4.png|800px|thumb|center|'''Figure | + | [[Image:DARPin4.png|800px|thumb|center|'''Figure 8:''' Flow cytometry of 6-well plate post incubation at 37 °C and 20% oxygen for an hour; MSCs: mesenchymal stem cells. mScarlet_DARPin929 denotes <partinfo>BBa_K3111201</partinfo>, DARPin929_mScarlet denotes <partinfo>BBa_K3111202</partinfo> and <i>T.maritima</i> + iLOV denotes <partinfo>BBa_K3111104</partinfo>. Gating determined by using control SK-BR-3 and MSC cells without any additives.]] |
| − | The flow cytometry data allowed precise quantification of the population of cells that has the DARPin929 bound on the surface. In the plots of Figure | + | The flow cytometry data allowed precise quantification of the population of cells that has the DARPin929 bound on the surface. In the plots of Figure 8, whatever is on the right-hand side of the red line shows binding and correspondingly whatever is on the left, no binding. Based on Figure 8 (a) & (d) we observe that DARPin929_mScarlet has better binding efficiency with 29.8% compared to mScarlet_DARPin929 with only 19.6%. Clearly it can be noticed, that there is no unspecific binding when no DARPin is present as in the cases of rTurboGFP and <i>T. maritima</i> encapsulin fused with iLOV as well as when the DARPin fusion proteins have no receptor to bind to as in the case of Figure 8 (b) and (e). |
Furthermore, we wanted to investigate the rate of internalisation by observing the change in binding percentage at different incubation times, therefore we conducted further experiments investigating different incubation times with our construct of interest (mScarlet_DARPin929 - <partinfo>BBa_K3111201</partinfo>). | Furthermore, we wanted to investigate the rate of internalisation by observing the change in binding percentage at different incubation times, therefore we conducted further experiments investigating different incubation times with our construct of interest (mScarlet_DARPin929 - <partinfo>BBa_K3111201</partinfo>). | ||
| − | [[Image:mS_DARPin5.png|800px|thumb|center|'''Figure | + | [[Image:mS_DARPin5.png|800px|thumb|center|'''Figure 9:''' Flow cytometric analysis of SKBR3 cell incubated with mScarlet_DARPin929 (3μM) post incubation at 37 °C and 2% oxygen to observe the pattern of binding at different incubation times. Gating determined by using control SK-BR-3 cells without any additives.]] |
| − | While we would expect binding to reduce over the span of the different incubation times, this was not observed from the flow cytometric analysis in Figure | + | While we would expect binding to reduce over the span of the different incubation times, this was not observed from the flow cytometric analysis in Figure 9. We could observe that over time the binding of mScarlet_DARPin929 is increasing. We speculate that internalisation happens simultaneously as it can be observed in the images of Figure 9 where the fluorescence is observed across the cytoplasm adjacent to the DAPI-stained nucleus. However, to obtain more accurate data regarding this, future experiments would involve lysing the cells to release the fluorescent hybrid DARPin929 protein. |
===Performance in encapsulin-based drug delivery system=== | ===Performance in encapsulin-based drug delivery system=== | ||
| + | |||
| + | For our final experiments, we tested whether DARPins retain their binding characterists when fused to an encapsulin. For this we used expressed encapsulin-DARPin fusion protein, packaged with miniSOG as described in part <partinfo>BBa_K3111502</partinfo>. | ||
| + | |||
| + | The cells were incubated with the Encapsulin-DARPin929-miniSOG drug delivery construct for 60 minutes at 37 °C and 20% oxygen. At the end of the experiment, the cells were visualised with EVOS FL microscope to observe uptake of the encapsulins. Once again, we used Mesenchymal Stem Cells (MSCs), which do not possess the HER2 receptor, for our control experiments. | ||
| + | |||
| + | [[Image:TmEncH_DARPin929_miniSOG3.png|500px|thumb|center|'''Figure 10:''' Confocal Microscopy image of MSCs and SK-BR-3 after 60-minute incubation; Scalebar: 200 μm.]] | ||
| + | |||
| + | From the confocal microscopy imaging in Figure 10, we could see that after 1-hour incubation green fluorescence from miniSOG was detected in both samples. Fluorescence was not expected for the MSCs, since from our previous experiments we have not observed any unspecific binding. We hypothesise that the concentration of construct that we loaded was too high, thus non-specific passive uptake was initiated. However, since significantly more fluorescence was observed in SK-BR-3 cells, we can deem that the system is indeed selective. | ||
| + | |||
| + | ===Conclusion=== | ||
| + | |||
| + | Throughout these experiments we have explored the properties and potential of DARPin929 as an HER2 binding peptide. We have showcased the ability of DARPin929 (i) to be expressed in bacterial chassis, (i) to be fused to either terminus of a protein and (iii) to retain its properties upon fusion on the outer surface of <i>Thermotoga maritima</i> encapsulin. We hope these experiments will give a basis for further use of DARPins as highly selective binding molecules. | ||
| + | |||
| + | ===References=== | ||
| + | |||
| + | [1] Plückthun A. Designed Ankyrin Repeat Proteins (DARPins): Binding Proteins for Research, Diagnostics, and Therapy. Annual Review of Pharmacology and Toxicology. 2015;55(1):489-511. | ||
| + | |||
| + | [2] Janeway CA, Medzhitov R. Innate Immune Recognition. Annu Rev Immunol. 1 de abril de 2002;20(1):197-216. | ||
| + | |||
| + | [3] Forrer P, Stumpp MT, Binz HK, Plückthun A. A novel strategy to design binding molecules harnessing the modular nature of repeat proteins. FEBS Lett. 27 de marzo de 2003;539(1):2-6. | ||
| + | |||
| + | [4] Binz HK, Stumpp MT, Forrer P, Amstutz P, Plückthun A. Designing Repeat Proteins: Well-expressed, Soluble and Stable Proteins from Combinatorial Libraries of Consensus Ankyrin Repeat Proteins. J Mol Biol. 12 de septiembre de 2003;332(2):489-503. | ||
| + | |||
| + | [5] Stumpp MT, Binz HK, Amstutz P. DARPins: A new generation of protein therapeutics. Drug Discov Today. 1 de agosto de 2008;13(15):695-701. | ||
| + | |||
| + | [6] Using Mimics to Get Around Antibodies’ Limitations [Internet]. The Scientist Magazine®. [cited on 26th June 2019]. Available at: https://www.the-scientist.com/lab-tools/using-mimics-to-get-around-antibodies-limitations-64264 | ||
| + | |||
| + | [7] Eggel A, Baumann MJ, Amstutz P, Stadler BM, Vogel M. DARPins as bispecific receptor antagonists analyzed for immunoglobulin E receptor blockage. J Mol Biol. octubre de 2009;393(3):598-607. | ||
| + | |||
| + | [8] DARPins: an eye-opening new biologic therapy [Internet]. [cited on 26th June 2019]. Available at: https://www.drugdevelopment-technology.com/comment/darpins-eye-opening-new-biologic-therapy/ | ||
| + | |||
| + | [9] 21. Camacho-Leal MP, Sciortino M, Cabodi S. ErbB2 Receptor in Breast Cancer: Implications in Cancer Cell Migration, Invasion and Resistance to Targeted Therapy, Breast Cancer - From Biology to Medicine. Phuc Van Pham, IntechOpen. | ||
| + | |||
| + | [10] Siegler E, Li S, Kim YJ, Wang P. Designed Ankyrin Repeat Proteins as Her2 Targeting Domains in Chimeric Antigen Receptor-Engineered T Cells. Hum Gene Ther [Internet]. 2017 Sep 1 [cited 2019 Sep 14];28(9):726–36. | ||
| + | |||
| + | [11] Steiner D, Forrer P, Plückthun A. Efficient Selection of DARPins with Sub-nanomolar Affinities using SRP Phage Display. J Mol Biol [Internet]. 2008 Oct 24 [cited 2019 Sep 14];382(5):1211–27. | ||
Latest revision as of 18:09, 30 September 2024
DARPin929
This part encodes DARPin929 – a binding protein specific to HER2 receptor, that is similar to an antibody in terms of efficiency and specificity. However, due to its stability and ability to be expressed in bacteria, this DARPin can be used in a wider variety of applications (1). The part has had its start and stop codons removed for flexible fusion to N or C terminus.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Contents
Usage and Biology
Designed Ankyrin Repeat Proteins (DARPins) are small (14-21 kDa) single-domain binding molecules which can bind to specific targets and subsequently trigger various molecular mechanisms (e.g. enzyme inhibition or protein anchoring) (2,3). They have been genetically engineered by Andreas Plückthun in 2003, as a means to manipulate the properties of natural ankyrin repeat proteins for therapeutic applications (4,5). New DARPins can be created via mutagenesis and phase display technologies, which enables the selection of highly stable, soluble, easily-expressed and efficient versions. This indicates that these binding molecules have the potential to circumvent the limitations of monoclonal antibodies and empower new therapeutic approaches (4). In short, they possess the following qualities making them ideal for therapeutic approaches:
- Due to their high specificity, stability (thermodynamic), and affinity, as well as their flexible architecture, DARPins have a rigid body-binding mode. They are stable in human blood serum and can be engineered so as not to contain T-cell epitopes (5).
- Their unique pharmacokinetic properties can be altered upon fusion with half-life extending molecules (5,7).
- They can be produced rapidly and cost-effectively (i.e. in E. coli) (as opposed to antibodies that are expensive, have a tedious manufacturing process and need to be humanized before administration) (8).
- Multi-specific DARPin constructs can be made by genetic fusion while retaining similar binding properties as single-domain DARPins, meaning that they can provide greater clinical benefit by overcoming the limitations of conventional therapeutic approaches, that typically target a single disease pathway (many diseases result from the dysregulation of multiple pathways). DARPin technology can be harnessed to rapidly generate many different “multi-DARPins” which enable targeting of multiple disease pathways (1,6,7).
- DARPins have been shown to be very stable and show less tendencies for aggregation, unlike single chain variable fragments since they do not possess cysteines.
DARPin929, in particular, has been one of the most frequently used to evaluate probes for molecular imaging. It particularly targets human epidermal growth factor 2 (HER2) whose overexpression is associated with breast cancer and gastroesophageal cancer (9). It has been shown to bind to its target with nanomolar affinities (10,11).
Functionalisation
BBa_K3111011 has been used along with BBa_K3111021 for initial investigation of binding to SK-BR-3 HER2+ breast adenocarcinoma cells. Moreover, it was fused with BBa_K3111003 and co-expressed with BBa_K3111032 to create the drug delivery vehicle containing cytotoxic cargo (BBa_K3111502).
Structural modelling of DARPin929 fusion to encapsulins
We suspected that fusing DARPin929 to the outer surfaces of T. maritima and M. xanthus encapsulins may result in steric clashes between encapsulin-DARPin dimers, thus impeding or preventing the assembly of the full encapsulin capsules. In order to assess the feasibility of fusing DARPin929 to either encapsulin, we created protein models using PyMol (GUI and manual edits), Discovery Studio (structure quality), GROMACS (energy minimisation of created dimers) and atomium (rebuilding the full encapsulin capsules).
Figures 1 and 2 show the full capsule models of T. maritima (Fig. 1) and M. xanthus (Fig. 2) encapsulins with DARPins bound to outer terminal of each monomer.
The clashes present in Figure 2 are an artefact of building the model and would not be seen in vivo. However, the overall space for the DARPins to resolve without clashes and rotate to bind HER2 is evidently bigger with the T. maritima encapsulin.
As a result we conclude that fusing DARPin929 to the T. maritima encapsulin is more readily feasible. We demonstrate this fusion in BBa_K3111501.
Experimental Results
Experiments showing the properties of BBa_K3111011 were carried out using composite parts BBa_K3111201, BBa_K3111202, BBa_K3111501, BBa_K3111502 and BBa_K3111503.
Fusion to BBa_K3111003
Confirmation of assembly
Using composite part BBa_K3111501 we investigated the ability of DARPin929 to be fused onto T. maritima encapsulin without hindering its assembly. We first demonstrated that this fusion protein would express and purify without cleavage by running it on an SDS gel (Figure 4). From this we could observe that the encapsulin became strongly insoluble after the addition of DARPin929, however, a small fraction of it could still be purified, as seen in lane 2 and 3 on the gel (~51 kDa). We did not observe any smaller bands (besides host cell protein at ~15 kDa), indicating that the DARPin was not cleaved off.
To observe assembly under non-reducing environment, we concentrated the purified sample from elution 2 and used it for Transmission Electron Microscopy imaging and non-reducing PAGE gel.
Figure 5 (a) shows the TEM image obtained. We could clearly observe assembled encapsulins thus concluding that although expression of the protein was mostly insoluble, the soluble fragment contained fully assembled encapsulins. The native PAGE gel observed in Figure 5 (b) let us confirm that the DARPin did not get cleaved-off during assembly, as we observed a significant increase in band size from the TmEnc lane to the TmEnc_DARPin929.
After these observations we decided that assembly was not very critically affected and moved to cargo loading in part BBa_K3111502.
Impact of DARPin fusion on cargo loading
Furthermore, we investigated the impact of fusing DARPin929 on cargo loading into the encapsulin by evaluating two different DARPin display strategies. First, BBa_K3111502 showed the loading of mini singlet oxyged generator (miniSOG) into an encapsulin which has a DARPin on every monomer, whereas, BBa_K3111503 had a mixture of encapsulin monomer with and without a DARPin.
Figure 6 shows the cargo loading capacity obtained from producing the multicomponent drug delivery platform using the aforementioned cloning strategies. Loading capacity for BBa_K3111502 was estimated to be 13.61+-0.36 molecules of miniSOG. In contrast, BBa_K3111503 was shown to be loading 8.19±0.21 miniSOG molecules per encapsulin. We hypothesise that this difference is caused by the surface display of DARPins slowing down the assembly of encapsulins and giving the targeting peptide present on miniSOG more time to form hydrophobic interactions to the inner shell of encapsulin monomers. Analysis on the methodology used to calculate the number of loaded cargo proteins can be found on BBa_K3111402 registry page.
Evaluation of binding
To successfully display a DARPin on the encapsulin surface, it was required to fuse it downstream of the encapsulin sequence. However, we could not find any recorded cases of fusing a DARPin on the C-terminus of another protein. We hyphotesised that C-terminal fusion could obstruct the binding site of the DARPin, therefore, we decided to test this with composite parts BBa_K3111201 and BBa_K3111202. These parts featured DARPin929 fused to the red fluorescent protein mScarlet via 6-aa flexible linker in two different orientations. We tested the constructs on HER2 overexpressing SK-BR-3 breast adenocarcinoma cells, to which DARPin929 can bind to with nanomolar affinities.
Our preliminary experiments (found on the composite part pages: BBa_K3111201 and BBa_K3111202) determined that the optimal conditions for the binding of DARPin929 were 37 °C and 3 μM with no significant difference observed between 2% or 20% oxygen. In order to validate the binding specificity of DARPins, we conducted further control mammalian cell culture experiments by applying the optimal conditions. These experiments were done to firstly make sure that proteins without fused DARPins would not get uptaken by the cells or bind to their surface. This was done by applying rTurboGFP at the same concentration. Secondly to observe the specificity of DARPin and subsequently our drug delivery platform we then applied the DARPin + mScarlet fusions, on Mesenchymal stem cells (MSCs) which do not express HER2 receptors. Additionally, we wanted to observe the interaction between T. maritima encapsulins and SK-BR-3 cells for future experiments. Thus we manufactured encapsulins without DARPins (BBa_K3111104) but surface displayed iLOV, a cyan fluorescent protein and applied it at the same concentration as the previous constructs.

From Figure 7, we observed no fluorescence thus no binding or uptake of both rTurboGFP and Thermotoga maritima encapsulin fused with iLOV. Thus, we confirm that in the absence of DARPins fusion proteins cannot bind or get internalised. Moreover, we observe no red fluorescence on the MSCs indicating the specificity of DARPin only for HER2 receptors, thus confirming the potential for targeted delivery platforms by using DARPin.
To obtain greater accuracy of binding percentage we proceeded with flow cytometry. Thus, we dislodged the cells from their growth wells using EDTA, while not affecting the binding of the DARPin and the HER2 receptor, and then used the sample for flow cytometry. We used the flow cytometer (BD AccuriTM C6 Plus) configured to observe the fluorescence within the mScarlet and CFP regions.

The flow cytometry data allowed precise quantification of the population of cells that has the DARPin929 bound on the surface. In the plots of Figure 8, whatever is on the right-hand side of the red line shows binding and correspondingly whatever is on the left, no binding. Based on Figure 8 (a) & (d) we observe that DARPin929_mScarlet has better binding efficiency with 29.8% compared to mScarlet_DARPin929 with only 19.6%. Clearly it can be noticed, that there is no unspecific binding when no DARPin is present as in the cases of rTurboGFP and T. maritima encapsulin fused with iLOV as well as when the DARPin fusion proteins have no receptor to bind to as in the case of Figure 8 (b) and (e).
Furthermore, we wanted to investigate the rate of internalisation by observing the change in binding percentage at different incubation times, therefore we conducted further experiments investigating different incubation times with our construct of interest (mScarlet_DARPin929 - BBa_K3111201).
While we would expect binding to reduce over the span of the different incubation times, this was not observed from the flow cytometric analysis in Figure 9. We could observe that over time the binding of mScarlet_DARPin929 is increasing. We speculate that internalisation happens simultaneously as it can be observed in the images of Figure 9 where the fluorescence is observed across the cytoplasm adjacent to the DAPI-stained nucleus. However, to obtain more accurate data regarding this, future experiments would involve lysing the cells to release the fluorescent hybrid DARPin929 protein.
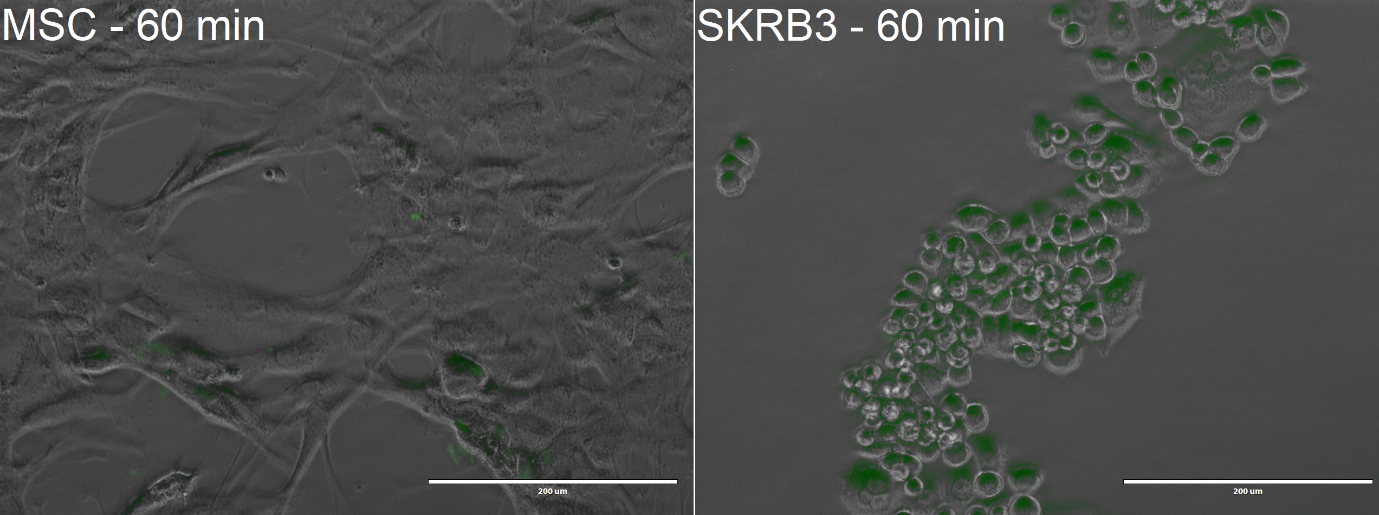
Performance in encapsulin-based drug delivery system
For our final experiments, we tested whether DARPins retain their binding characterists when fused to an encapsulin. For this we used expressed encapsulin-DARPin fusion protein, packaged with miniSOG as described in part BBa_K3111502.
The cells were incubated with the Encapsulin-DARPin929-miniSOG drug delivery construct for 60 minutes at 37 °C and 20% oxygen. At the end of the experiment, the cells were visualised with EVOS FL microscope to observe uptake of the encapsulins. Once again, we used Mesenchymal Stem Cells (MSCs), which do not possess the HER2 receptor, for our control experiments.
From the confocal microscopy imaging in Figure 10, we could see that after 1-hour incubation green fluorescence from miniSOG was detected in both samples. Fluorescence was not expected for the MSCs, since from our previous experiments we have not observed any unspecific binding. We hypothesise that the concentration of construct that we loaded was too high, thus non-specific passive uptake was initiated. However, since significantly more fluorescence was observed in SK-BR-3 cells, we can deem that the system is indeed selective.
Conclusion
Throughout these experiments we have explored the properties and potential of DARPin929 as an HER2 binding peptide. We have showcased the ability of DARPin929 (i) to be expressed in bacterial chassis, (i) to be fused to either terminus of a protein and (iii) to retain its properties upon fusion on the outer surface of Thermotoga maritima encapsulin. We hope these experiments will give a basis for further use of DARPins as highly selective binding molecules.
References
[1] Plückthun A. Designed Ankyrin Repeat Proteins (DARPins): Binding Proteins for Research, Diagnostics, and Therapy. Annual Review of Pharmacology and Toxicology. 2015;55(1):489-511.
[2] Janeway CA, Medzhitov R. Innate Immune Recognition. Annu Rev Immunol. 1 de abril de 2002;20(1):197-216.
[3] Forrer P, Stumpp MT, Binz HK, Plückthun A. A novel strategy to design binding molecules harnessing the modular nature of repeat proteins. FEBS Lett. 27 de marzo de 2003;539(1):2-6.
[4] Binz HK, Stumpp MT, Forrer P, Amstutz P, Plückthun A. Designing Repeat Proteins: Well-expressed, Soluble and Stable Proteins from Combinatorial Libraries of Consensus Ankyrin Repeat Proteins. J Mol Biol. 12 de septiembre de 2003;332(2):489-503.
[5] Stumpp MT, Binz HK, Amstutz P. DARPins: A new generation of protein therapeutics. Drug Discov Today. 1 de agosto de 2008;13(15):695-701.
[6] Using Mimics to Get Around Antibodies’ Limitations [Internet]. The Scientist Magazine®. [cited on 26th June 2019]. Available at: https://www.the-scientist.com/lab-tools/using-mimics-to-get-around-antibodies-limitations-64264
[7] Eggel A, Baumann MJ, Amstutz P, Stadler BM, Vogel M. DARPins as bispecific receptor antagonists analyzed for immunoglobulin E receptor blockage. J Mol Biol. octubre de 2009;393(3):598-607.
[8] DARPins: an eye-opening new biologic therapy [Internet]. [cited on 26th June 2019]. Available at: https://www.drugdevelopment-technology.com/comment/darpins-eye-opening-new-biologic-therapy/
[9] 21. Camacho-Leal MP, Sciortino M, Cabodi S. ErbB2 Receptor in Breast Cancer: Implications in Cancer Cell Migration, Invasion and Resistance to Targeted Therapy, Breast Cancer - From Biology to Medicine. Phuc Van Pham, IntechOpen.
[10] Siegler E, Li S, Kim YJ, Wang P. Designed Ankyrin Repeat Proteins as Her2 Targeting Domains in Chimeric Antigen Receptor-Engineered T Cells. Hum Gene Ther [Internet]. 2017 Sep 1 [cited 2019 Sep 14];28(9):726–36.
[11] Steiner D, Forrer P, Plückthun A. Efficient Selection of DARPins with Sub-nanomolar Affinities using SRP Phage Display. J Mol Biol [Internet]. 2008 Oct 24 [cited 2019 Sep 14];382(5):1211–27.