Difference between revisions of "Part:BBa K3056000"
| (51 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
<partinfo>BBa_K3056000 short</partinfo> | <partinfo>BBa_K3056000 short</partinfo> | ||
<br/> | <br/> | ||
| − | < | + | <font size="4"> |
| − | '''α -THC Antibody Conjugate to mNG:''' | + | '''α -THC Antibody Conjugate to mNG:'''<br/> |
Validation of expression, fluorescence, and binding ability to Δ9-tetrahydrocannabinol (henceforth, THC). | Validation of expression, fluorescence, and binding ability to Δ9-tetrahydrocannabinol (henceforth, THC). | ||
<br/> | <br/> | ||
<br/> | <br/> | ||
| − | '''Components''' | + | '''Components'''<br/> |
This part consists of a periplasmic localization signal (PLS), and an anti-Δ9-tetrahydrocannabinol ScFv linked to mNG at the C-terminal. Note that this part contains a stop codon. | This part consists of a periplasmic localization signal (PLS), and an anti-Δ9-tetrahydrocannabinol ScFv linked to mNG at the C-terminal. Note that this part contains a stop codon. | ||
<br/> | <br/> | ||
| − | + | ---- | |
| − | '''Summary''' | + | '''Summary'''<br/> |
A fluorescently labelled anti-THC antibody was successfully produced in E. coli. The antibody fragment was able to bind THC soaked lipophilic membranes, with a sensitivity of 0.1 mg/mL. Future experiments may aim to increase the sensitivity of the THC assay by changing the fluorescent tags or testing a larger library of lipophilic membranes. | A fluorescently labelled anti-THC antibody was successfully produced in E. coli. The antibody fragment was able to bind THC soaked lipophilic membranes, with a sensitivity of 0.1 mg/mL. Future experiments may aim to increase the sensitivity of the THC assay by changing the fluorescent tags or testing a larger library of lipophilic membranes. | ||
<br/> | <br/> | ||
| − | + | ---- | |
| − | '''Background''' | + | '''Background'''<br/> |
Here we designed a fluorescent anti-THC antibody, optimized for E. coli expression. Recombinant antibody expression in E. coli is notoriously challenging, as typical IgG proteins require post-translational modifications. However, Recombinant expression of antibodies can be made possible by truncating antibody fragment ('''Fig. 1''').<br> | Here we designed a fluorescent anti-THC antibody, optimized for E. coli expression. Recombinant antibody expression in E. coli is notoriously challenging, as typical IgG proteins require post-translational modifications. However, Recombinant expression of antibodies can be made possible by truncating antibody fragment ('''Fig. 1''').<br> | ||
The protein sequence for the anti-THC fragment (ScFv) had been previously characterized and is optimized for E. coli expression systems ('''1'''). Hence, this part is codon optimized for E. coli. The protein sequence also contains a periplasmic localization signal (PLS), encoded by the OmpA sequence. Periplasmic localization provides an oxidizing environment for disulfide bonds in the antibody to form. to further improve disulfide bond formation, the protein may be expressed in an engineered cell line with an oxidizing cytoplasmic environment (Ex: Rosetta gami2). | The protein sequence for the anti-THC fragment (ScFv) had been previously characterized and is optimized for E. coli expression systems ('''1'''). Hence, this part is codon optimized for E. coli. The protein sequence also contains a periplasmic localization signal (PLS), encoded by the OmpA sequence. Periplasmic localization provides an oxidizing environment for disulfide bonds in the antibody to form. to further improve disulfide bond formation, the protein may be expressed in an engineered cell line with an oxidizing cytoplasmic environment (Ex: Rosetta gami2). | ||
<br/> | <br/> | ||
| − | [[File:T--Queens_Canada--Antibody_Trunc.jpg|thumb|420px|center|<b>'''Figure 1.'''</b>The modification of an IgG protein to a Fab and ScFv, which can be expressed in E. coli. In the case of the anti-THC antibody, the protein was conjugated to a fluorescent protein at the C-terminal.]] <br/> | + | [[File:T--Queens_Canada--Antibody_Trunc.jpg|thumb|420px|center|<b>'''Figure 1.'''</b> The modification of an IgG protein to a Fab and ScFv, which can be expressed in E. coli. In the case of the anti-THC antibody, the protein was conjugated to a fluorescent protein at the C-terminal.]]<br/> |
| − | + | ---- | |
| − | + | '''Design'''<br/> | |
| − | + | ||
| − | '''Design''' | + | |
The structure of the anti-THC ScFv has not been determined; hence, we modelled the ScFv to determine optimal linkage to a fluorescent protein. ABodyBuilder predicted the structure of the anti-THC ScFv, based on template selection, orientation prediction, complementary-determining region (CDR) loop modeling, and side chain prediction ('''2'''). The root-mean-square deviation (RMSD) for the predicted heavy and light chain model are 1.00, and 0.88, respectively, indicating high model confidence ('''Fig. 2'''). Moreover, the predicted CDR regions determined by ABodyBuilder agree with the previously predicted CDRs by the researchers who characterized the antibody ('''1'''). Additionally, the model indicated that the N and C-terminal of the light chain were too close to the binding site; however, the C-terminal of the heavy chain was suitable for linkage to a fluorescent protein ('''Fig. 3'''). Therefore, the fluorescent protein was linked to the ScFv on the C-terminal of the heavy chain. | The structure of the anti-THC ScFv has not been determined; hence, we modelled the ScFv to determine optimal linkage to a fluorescent protein. ABodyBuilder predicted the structure of the anti-THC ScFv, based on template selection, orientation prediction, complementary-determining region (CDR) loop modeling, and side chain prediction ('''2'''). The root-mean-square deviation (RMSD) for the predicted heavy and light chain model are 1.00, and 0.88, respectively, indicating high model confidence ('''Fig. 2'''). Moreover, the predicted CDR regions determined by ABodyBuilder agree with the previously predicted CDRs by the researchers who characterized the antibody ('''1'''). Additionally, the model indicated that the N and C-terminal of the light chain were too close to the binding site; however, the C-terminal of the heavy chain was suitable for linkage to a fluorescent protein ('''Fig. 3'''). Therefore, the fluorescent protein was linked to the ScFv on the C-terminal of the heavy chain. | ||
<br/> | <br/> | ||
<br/> | <br/> | ||
| − | [[File:T--Queens_Canada--ScFv_mNG_confidence.jpg|thumb|left|500px|<b>'''Figure 2.'''</b>Confidence in the model obtained by ABodyBuilder. Note that the confidence score of the heavy chain (VH) and light chain (LH) are 1.00, and 0.88, respectively.]] | + | [[File:T--Queens_Canada--ScFv_mNG_confidence.jpg|thumb|left|500px|<b>'''Figure 2.'''</b> Confidence in the model obtained by ABodyBuilder. Note that the confidence score of the heavy chain (VH) and light chain (LH) are 1.00, and 0.88, respectively.]] |
| − | [[File:T--Queens_Canada--ScFv_mNG_2.jpg|thumb|175px|center|<b>'''Figure 3.'''</b>The predicted structure of the ScFv-mNG conjugate. mNG was only suitable to linkage by the c-terminal of the heavy chain.]] | + | [[File:T--Queens_Canada--ScFv_mNG_2.jpg|thumb|175px|center|<b>'''Figure 3.'''</b> The predicted structure of the ScFv-mNG conjugate. mNG was only suitable to linkage by the c-terminal of the heavy chain.]] |
| + | <br/> | ||
| + | ---- | ||
| + | [[File:T--Queens_Canada--ScFv-mNG_Digest.jpg|thumb|left|400px|<b>'''Figure 4.'''</b> Test digest of ScFv-mNG integration into the pET24d vector. The band above 1.5 kb indicates successful integration of the 1536 bp biobrick.]] | ||
<br/> | <br/> | ||
| − | |||
<br/> | <br/> | ||
'''Cloning''' <br/> | '''Cloning''' <br/> | ||
| − | The biobrick was ordered from IDT, digested, and ligated into pET24d. pET24d was chosen as a vector due to the availability of the T7 promoter. | + | The biobrick was ordered from IDT, digested, and ligated into pET24d ('''Fig. 4'''). pET24d was chosen as a vector due to the availability of the T7 promoter. |
<br/> | <br/> | ||
<br/> | <br/> | ||
| + | |||
| + | [[File:T--Queens_Canada--ScFv_expression.jpg|thumb|left|400px|<b>'''Figure 5.'''</b> Nickel column purification of ScFv-mNG and ScFv-EGFP. Both proteins were successfully purified from BL21 cells, as indicated by the major band at 56 kDa.]] | ||
| + | <br/> | ||
| + | <br/> | ||
| + | <br/> | ||
| + | <br/> | ||
| + | <br/> | ||
| + | <br/> | ||
| + | <br/> | ||
| + | <br/> | ||
| + | <br/> | ||
| + | <br/> | ||
| + | <br/> | ||
| + | |||
'''Expression'''<br/> | '''Expression'''<br/> | ||
| − | The biobrick was expressed in E. coli Rosetta gami2, as this cell line contains an oxidizing cytoplasmic environment ideal for the disulfide bond formations within the antibody ('''Fig. | + | The biobrick was expressed in E. coli Rosetta gami2, as this cell line contains an oxidizing cytoplasmic environment ideal for the disulfide bond formations within the antibody ('''Fig. 5'''). Following a nickle affinity column purification, the ScFv was observed at 56 kDa. Note that another biobrick (BBa_K3056001) was designed, with EGFP as the fluorescent protein, rather than mNG, to compare fluorescent intensities. |
<br/> | <br/> | ||
<br/> | <br/> | ||
<br/> | <br/> | ||
| − | |||
<br/><br/> | <br/><br/> | ||
<br/><br/> | <br/><br/> | ||
| Line 52: | Line 65: | ||
<br/> | <br/> | ||
<br/> | <br/> | ||
| + | ---- | ||
| + | '''THC Binding''' | ||
<br/> | <br/> | ||
| + | [[File:T--Queens_Canada--SpotAssay.jpg|thumb|left|300 px|<b>'''Figure 6.'''</b> THC membrane assay using purified ScFv-EGFP and ScFv-mNG. Membranes were saturated with 10 mg/mL of THC, washed, and probed with the purified ScFv. Only ScFv-mNG gave a positive signal in the assay]] | ||
| + | The purified EGFP and mNG linked ScFv were spot tested on a membrane assay, where only the ScFv was able to bind the THC soaked membrane ('''Fig. 6'''). To test the binding, lipophilic membranes were saturated with 10 mg/mL of THC, washed 10x with phosphate buffered saline (1% Tween-20), incubated with ScFv-EGFP, or ScFv-mNG, washed 3x with phosphate buffered saline (1% Tween-20), and imaged in Azure Biosystems 600. The excitation and emission wavelengths were 395 nm, and 509 nm, respectively. | ||
<br/> | <br/> | ||
<br/> | <br/> | ||
<br/> | <br/> | ||
| − | |||
<br/> | <br/> | ||
| − | |||
<br/> | <br/> | ||
| − | |||
| − | |||
<br/> | <br/> | ||
<br/> | <br/> | ||
| − | '''THC Binding Limit''' <br/> | + | <br/> |
| + | ---- | ||
| + | '''THC Binding Limit''' | ||
| + | <br/> | ||
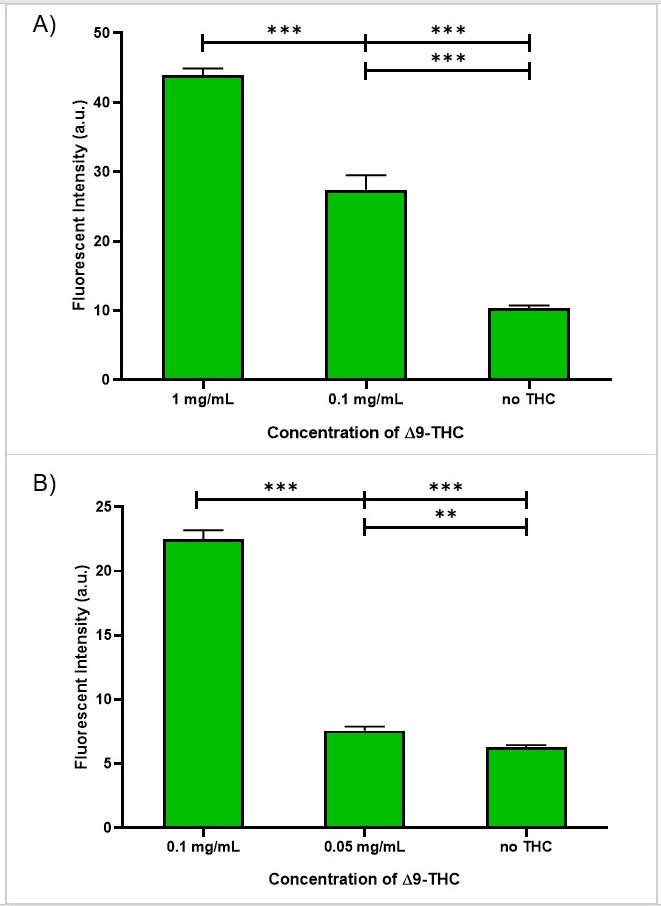
We further examined the detection limit of the anti-THC ScFv-mNG via the previously described membrane assay. The antibody signal was distinguishable from the background down to 0.1 mg/mL of THC ('''Fig. 7.''') At 0.1 mg/mL, the fluorescent intensity was ~3x as bright as the CTRL, which did not contain any THC ('''Fig. 8'''). | We further examined the detection limit of the anti-THC ScFv-mNG via the previously described membrane assay. The antibody signal was distinguishable from the background down to 0.1 mg/mL of THC ('''Fig. 7.''') At 0.1 mg/mL, the fluorescent intensity was ~3x as bright as the CTRL, which did not contain any THC ('''Fig. 8'''). | ||
| − | [[File:T--Queens_Canada--MembraneSens.jpg|thumb|left| | + | [[File:T--Queens_Canada--MembraneSens.jpg|thumb|left|475 px|<b>'''Figure 7.'''</b> THC membrane assay using ScFv-mNG and varying concentrations of THC. A) The signal in 0.1 mg/mL is distinguishable from the control (CTRL), indicating that the sensitivity is below 0.1 mg/mL. B) The signal in 0.05 mg/mL is not distinguishable from the control (CTRL). Therefore, the detection limit is between 0.1 and 0.05 mg/mL.]] |
| − | [[File:T--Queens_Canada--MembraneSensPlot.jpg|thumb|center|400 px|<b>'''Figure | + | [[File:T--Queens_Canada--MembraneSensPlot.jpg|thumb|center|400 px|<b>'''Figure 8.'''</b> Fluorescent intensity of the membrane assay using ScFv-mNG and varying concentrations of THC. A) The signal in 0.1 mg/mL is distinguishable from the control (CTRL), indicating that the sensitivity is below 0.1 mg/mL. B) The signal in 0.05 mg/mL is not distinguishable from the control (CTRL). Therefore, the detection limit is between 0.1 and 0.05 mg/mL..]] |
| − | + | ---- | |
<br/> | <br/> | ||
| + | '''Future Directions''' | ||
| + | The recombinant antibody was able to detect THC on a lipophilic membrane down to a concentration of 0.1 mg/mL of THC. However, it seems that this assay is limited by the lipophilic membranes. Our team used PIG® Oil-Only Absorbent Mats, which are designed for cleaning up oil spills and hence are not manufactured uniformly. Future studies may aim at testing a large library of membranes for this test, in order to absorb THC uniformly, and ultimately increase the sensitivity of this assay.<br/> | ||
| + | ---- | ||
'''Protein Sequence''' <br/> | '''Protein Sequence''' <br/> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<span style="color:purple;">MKKTAIAIAVALAGFATVAQA</span> | <span style="color:purple;">MKKTAIAIAVALAGFATVAQA</span> | ||
<span style="color:blue;">QVQLQESGPGLVKPSETLSLTCTVSGGSISSG<br/>YYWGWIRQPPGKGLEWIGSIYHSGSTYYNPSLKSRVTISVDTSKNQFSLKLSS<br/>VTAADTAVYYCARGSAKRAVKWGQGTLVTVSSGGGGSGGGGSGGSALQTVVTQ<br/>EPSFSVSPGGTVTLTCGLSSGSVSTSYYPSWYQQTPGQAPRTLIYSTNTRSSG<br/>VPDRFSGSILGNKAALTITGAQADDESDYYCVLYMGSGVVFGGGTKLTVLG</span> | <span style="color:blue;">QVQLQESGPGLVKPSETLSLTCTVSGGSISSG<br/>YYWGWIRQPPGKGLEWIGSIYHSGSTYYNPSLKSRVTISVDTSKNQFSLKLSS<br/>VTAADTAVYYCARGSAKRAVKWGQGTLVTVSSGGGGSGGGGSGGSALQTVVTQ<br/>EPSFSVSPGGTVTLTCGLSSGSVSTSYYPSWYQQTPGQAPRTLIYSTNTRSSG<br/>VPDRFSGSILGNKAALTITGAQADDESDYYCVLYMGSGVVFGGGTKLTVLG</span> | ||
<span style="color:red;">GSGSGS<br/></span> | <span style="color:red;">GSGSGS<br/></span> | ||
<span style="color:green;">MVSKGEEDNMASLPATHELHIFGSINGVDFDMVGQGTGNPNDGYEELN<br/>LKSTKGDLQFSPWILVPHIGYGFHQYLPYPDGMSPFQAAMVDGSGYQVHRTMQF<br/>EDGASLTVNYRYTYEGSHIKGEAQVKGTGFPADGPVMTNSLTAADWCRSKKTY<br/>PNDKTIISTFKWSYTTGNGKRYRSTARTTYTFAKPMAANYLKNQPMYVFRKTE<br/>LKHSKTELNFKEWQKAFTDVMGMDELYKENLYFQG</span> | <span style="color:green;">MVSKGEEDNMASLPATHELHIFGSINGVDFDMVGQGTGNPNDGYEELN<br/>LKSTKGDLQFSPWILVPHIGYGFHQYLPYPDGMSPFQAAMVDGSGYQVHRTMQF<br/>EDGASLTVNYRYTYEGSHIKGEAQVKGTGFPADGPVMTNSLTAADWCRSKKTY<br/>PNDKTIISTFKWSYTTGNGKRYRSTARTTYTFAKPMAANYLKNQPMYVFRKTE<br/>LKHSKTELNFKEWQKAFTDVMGMDELYKENLYFQG</span> | ||
| − | |||
<br/> | <br/> | ||
Purple: OmpA (PLS) | Purple: OmpA (PLS) | ||
| Line 98: | Line 103: | ||
Green: mNG | Green: mNG | ||
<br/> | <br/> | ||
| − | + | ---- | |
'''References''' <br/> | '''References''' <br/> | ||
1. Brennan, J. (2005) The production of recombinant single chain antibody fragments for the detection of illicit drug residues. doctoral thesis, Dublin City University, [online] http://doras.dcu.ie/17319/ (Accessed March 12, 2019) | 1. Brennan, J. (2005) The production of recombinant single chain antibody fragments for the detection of illicit drug residues. doctoral thesis, Dublin City University, [online] http://doras.dcu.ie/17319/ (Accessed March 12, 2019) | ||
| + | <br/> | ||
2. ABodyBuilder: Automated antibody structure prediction with data–driven accuracy estimation: mAbs: Vol 8, No 7 [online] https://www.tandfonline.com/doi/full/10.1080/19420862.2016.1205773?scroll=top&needAccess=true (Accessed October 9, 2019) | 2. ABodyBuilder: Automated antibody structure prediction with data–driven accuracy estimation: mAbs: Vol 8, No 7 [online] https://www.tandfonline.com/doi/full/10.1080/19420862.2016.1205773?scroll=top&needAccess=true (Accessed October 9, 2019) | ||
Latest revision as of 19:15, 11 October 2019
Anti-THC antibody fragment (ScFv) linked to mNG at the C-terminal
α -THC Antibody Conjugate to mNG:
Validation of expression, fluorescence, and binding ability to Δ9-tetrahydrocannabinol (henceforth, THC).
Components
This part consists of a periplasmic localization signal (PLS), and an anti-Δ9-tetrahydrocannabinol ScFv linked to mNG at the C-terminal. Note that this part contains a stop codon.
Summary
A fluorescently labelled anti-THC antibody was successfully produced in E. coli. The antibody fragment was able to bind THC soaked lipophilic membranes, with a sensitivity of 0.1 mg/mL. Future experiments may aim to increase the sensitivity of the THC assay by changing the fluorescent tags or testing a larger library of lipophilic membranes.
Background
Here we designed a fluorescent anti-THC antibody, optimized for E. coli expression. Recombinant antibody expression in E. coli is notoriously challenging, as typical IgG proteins require post-translational modifications. However, Recombinant expression of antibodies can be made possible by truncating antibody fragment (Fig. 1).
The protein sequence for the anti-THC fragment (ScFv) had been previously characterized and is optimized for E. coli expression systems (1). Hence, this part is codon optimized for E. coli. The protein sequence also contains a periplasmic localization signal (PLS), encoded by the OmpA sequence. Periplasmic localization provides an oxidizing environment for disulfide bonds in the antibody to form. to further improve disulfide bond formation, the protein may be expressed in an engineered cell line with an oxidizing cytoplasmic environment (Ex: Rosetta gami2).
Design
The structure of the anti-THC ScFv has not been determined; hence, we modelled the ScFv to determine optimal linkage to a fluorescent protein. ABodyBuilder predicted the structure of the anti-THC ScFv, based on template selection, orientation prediction, complementary-determining region (CDR) loop modeling, and side chain prediction (2). The root-mean-square deviation (RMSD) for the predicted heavy and light chain model are 1.00, and 0.88, respectively, indicating high model confidence (Fig. 2). Moreover, the predicted CDR regions determined by ABodyBuilder agree with the previously predicted CDRs by the researchers who characterized the antibody (1). Additionally, the model indicated that the N and C-terminal of the light chain were too close to the binding site; however, the C-terminal of the heavy chain was suitable for linkage to a fluorescent protein (Fig. 3). Therefore, the fluorescent protein was linked to the ScFv on the C-terminal of the heavy chain.
Cloning
The biobrick was ordered from IDT, digested, and ligated into pET24d (Fig. 4). pET24d was chosen as a vector due to the availability of the T7 promoter.
Expression
The biobrick was expressed in E. coli Rosetta gami2, as this cell line contains an oxidizing cytoplasmic environment ideal for the disulfide bond formations within the antibody (Fig. 5). Following a nickle affinity column purification, the ScFv was observed at 56 kDa. Note that another biobrick (BBa_K3056001) was designed, with EGFP as the fluorescent protein, rather than mNG, to compare fluorescent intensities.
THC Binding
The purified EGFP and mNG linked ScFv were spot tested on a membrane assay, where only the ScFv was able to bind the THC soaked membrane (Fig. 6). To test the binding, lipophilic membranes were saturated with 10 mg/mL of THC, washed 10x with phosphate buffered saline (1% Tween-20), incubated with ScFv-EGFP, or ScFv-mNG, washed 3x with phosphate buffered saline (1% Tween-20), and imaged in Azure Biosystems 600. The excitation and emission wavelengths were 395 nm, and 509 nm, respectively.
THC Binding Limit
We further examined the detection limit of the anti-THC ScFv-mNG via the previously described membrane assay. The antibody signal was distinguishable from the background down to 0.1 mg/mL of THC (Fig. 7.) At 0.1 mg/mL, the fluorescent intensity was ~3x as bright as the CTRL, which did not contain any THC (Fig. 8).

Future Directions
The recombinant antibody was able to detect THC on a lipophilic membrane down to a concentration of 0.1 mg/mL of THC. However, it seems that this assay is limited by the lipophilic membranes. Our team used PIG® Oil-Only Absorbent Mats, which are designed for cleaning up oil spills and hence are not manufactured uniformly. Future studies may aim at testing a large library of membranes for this test, in order to absorb THC uniformly, and ultimately increase the sensitivity of this assay.
Protein Sequence
MKKTAIAIAVALAGFATVAQA
QVQLQESGPGLVKPSETLSLTCTVSGGSISSG
YYWGWIRQPPGKGLEWIGSIYHSGSTYYNPSLKSRVTISVDTSKNQFSLKLSS
VTAADTAVYYCARGSAKRAVKWGQGTLVTVSSGGGGSGGGGSGGSALQTVVTQ
EPSFSVSPGGTVTLTCGLSSGSVSTSYYPSWYQQTPGQAPRTLIYSTNTRSSG
VPDRFSGSILGNKAALTITGAQADDESDYYCVLYMGSGVVFGGGTKLTVLG
GSGSGS
MVSKGEEDNMASLPATHELHIFGSINGVDFDMVGQGTGNPNDGYEELN
LKSTKGDLQFSPWILVPHIGYGFHQYLPYPDGMSPFQAAMVDGSGYQVHRTMQF
EDGASLTVNYRYTYEGSHIKGEAQVKGTGFPADGPVMTNSLTAADWCRSKKTY
PNDKTIISTFKWSYTTGNGKRYRSTARTTYTFAKPMAANYLKNQPMYVFRKTE
LKHSKTELNFKEWQKAFTDVMGMDELYKENLYFQG
Purple: OmpA (PLS)
Blue: anti-THC ScFv
Red: Linker
Green: mNG
References
1. Brennan, J. (2005) The production of recombinant single chain antibody fragments for the detection of illicit drug residues. doctoral thesis, Dublin City University, [online] http://doras.dcu.ie/17319/ (Accessed March 12, 2019)
2. ABodyBuilder: Automated antibody structure prediction with data–driven accuracy estimation: mAbs: Vol 8, No 7 [online] https://www.tandfonline.com/doi/full/10.1080/19420862.2016.1205773?scroll=top&needAccess=true (Accessed October 9, 2019)






