Difference between revisions of "Part:BBa K3113103"
Theresakeil (Talk | contribs) |
|||
| (3 intermediate revisions by the same user not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K3113103 short</partinfo> | <partinfo>BBa_K3113103 short</partinfo> | ||
| − | This sequence codes for a coiled-coil structure which interacts with the coiled-coil P9SN. We use this construct to load RNA binding proteins into virus-like | + | This sequence codes for a coiled-coil structure which interacts with the coiled-coil peptide P9SN. We use this construct to load RNA binding proteins into virus-like particles. "Protein coiled-coil (CC) dimers serve as building blocks for modular de novo design of polyhedral protein cages that efficiently self-assemble in vitro and in vivo" (Ljubetic, A., et al. (2017)). |
| + | |||
| + | <h2>Usage</h2> | ||
| + | |||
| + | To increase the modularity of our system we decided to test coiled-coil proteins. These coiled coils mimic the interactions of DNA helix-helix interactions. The coiled-coil protein interaction allows the loading of more than one protein or larger proteins. We are using these helical structures to load RNA binding proteins into exosomes or virus-like-particles. | ||
| + | |||
| + | <h2>Biology</h2> | ||
| + | |||
| + | "We describe a system analogous to designed DNA nanostructures in which protein coiled-coil (CC) dimers serve as building blocks for modular de novo design of polyhedral protein cages that efficiently self-assemble in vitro and in vivo."<ref>Ljubetič, A., et al. (2017). "Design of coiled-coil protein-origami cages that self-assemble in vitro and in vivo." Nature Biotechnology 35: 1094.</ref> | ||
| + | |||
| + | <h2>Characterization</h2> | ||
| + | |||
| + | <h3>Western Blot</h3> | ||
| + | <html> | ||
| + | <figure class="figure"> | ||
| + | <img src="https://2019.igem.org/wiki/images/4/4f/T--Munich--WesternBlot_CC_test.png" width="50%" class="figure-img img-fluid rounded" alt=" "> | ||
| + | <figcaption style="font-size: 80%"> | ||
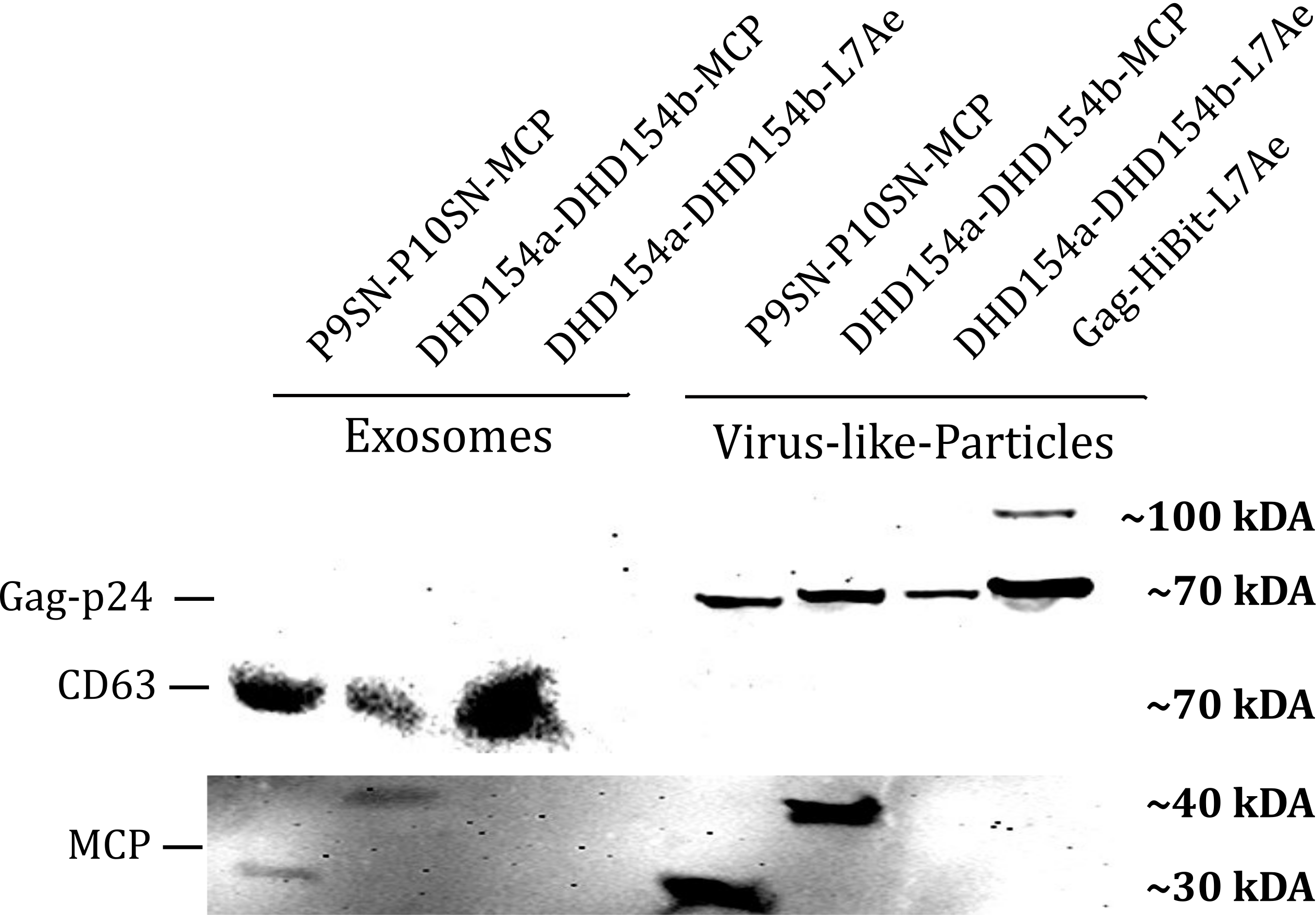
| + | <b>Figure 1: </b>The presence of vesicular components modularly composed with coiled-coils and directly fused could be proven by western blotting. Top left) the exosomal marker CD63 can be visualized with primary anti-CD63 mouse-antibody and secondary anti-mouse antibody - horseradish peroxidase (HRP) fusion. CD63 does not run as a tight band on the blot because of glycosylation patterns and its nature as a membrane protein. Top right) Gag-protein is determined at around 70 kDa. The fusion construct Gag-HiBiT-L7Ae shows some degradation corresponding to the molecular weight of L7Ae cleavage. Bottom) MCP RNA-binding proteins can be shown with anti-MCP antibodies. | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | </html> | ||
| + | |||
| + | <h3>qPCR</h3> | ||
| + | |||
| + | Using a coiled-coil system (Figure 7) allowed us to successfully load the two different RNA binding proteins (RBPs) L7Ae or MCP into exosomes to export FLuc mRNA from HEK293T cells. We used the published parallel heterodimer pair P9SN:P10SN (Ljubetič et al. 2017), where P9SN was fused to the C-terminus of the exosome marker protein CD63, and P10SN N-terminal to the RNA binding proteins L7Ae or MCP. The export efficiencies of FLuc mRNA cargo measured by qPCR proved that coiled-coil mediated targeting of the RBPs L7Ae and MCP into exosomes worked. Exosomes formed from CD63-P9SN and loaded with P10SN-L7Ae or -MCP could export about 800 and 400 transcripts per cell, respectively. This also shows that the two RBPs have different export capabilities, with L7Ae exporting twice as many transcripts per cell compared to MCP. | ||
| + | |||
| + | <html> | ||
| + | <figure class="figure"> | ||
| + | <img src="https://2019.igem.org/wiki/images/6/61/T--Munich--Modularity_of_engineering.png" width="50%" class="figure-img img-fluid rounded" style="display: float" alt="placeholder"> | ||
| + | <figcaption style="font-size: 80%"> | ||
| + | <b>Figure 7: Vesicles can be modularly assembled via coiled-coils.</b> Two different RNA binding proteins (RBP) were fused to the coiled-coil (CC) peptide P10SN and loaded into exosomes displaying the complementary part P9SN. qPCR analysis demonstrated specific export of target FLuc mRNA and a higher efficiency for L7Ae compared to MCP. Quantification was done via standard curve measurements and assuming a confluent well with 350.000 HEK293T cells/cm2 at the time of harvesting. n = 2 | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | </html> | ||
| + | |||
| − | |||
| − | |||
<!-- --> | <!-- --> | ||
| Line 12: | Line 44: | ||
<partinfo>BBa_K3113103 SequenceAndFeatures</partinfo> | <partinfo>BBa_K3113103 SequenceAndFeatures</partinfo> | ||
| + | <h2>References</h2> | ||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
Latest revision as of 01:50, 22 October 2019
P10SN
This sequence codes for a coiled-coil structure which interacts with the coiled-coil peptide P9SN. We use this construct to load RNA binding proteins into virus-like particles. "Protein coiled-coil (CC) dimers serve as building blocks for modular de novo design of polyhedral protein cages that efficiently self-assemble in vitro and in vivo" (Ljubetic, A., et al. (2017)).
Usage
To increase the modularity of our system we decided to test coiled-coil proteins. These coiled coils mimic the interactions of DNA helix-helix interactions. The coiled-coil protein interaction allows the loading of more than one protein or larger proteins. We are using these helical structures to load RNA binding proteins into exosomes or virus-like-particles.
Biology
"We describe a system analogous to designed DNA nanostructures in which protein coiled-coil (CC) dimers serve as building blocks for modular de novo design of polyhedral protein cages that efficiently self-assemble in vitro and in vivo."[1]
Characterization
Western Blot
qPCR
Using a coiled-coil system (Figure 7) allowed us to successfully load the two different RNA binding proteins (RBPs) L7Ae or MCP into exosomes to export FLuc mRNA from HEK293T cells. We used the published parallel heterodimer pair P9SN:P10SN (Ljubetič et al. 2017), where P9SN was fused to the C-terminus of the exosome marker protein CD63, and P10SN N-terminal to the RNA binding proteins L7Ae or MCP. The export efficiencies of FLuc mRNA cargo measured by qPCR proved that coiled-coil mediated targeting of the RBPs L7Ae and MCP into exosomes worked. Exosomes formed from CD63-P9SN and loaded with P10SN-L7Ae or -MCP could export about 800 and 400 transcripts per cell, respectively. This also shows that the two RBPs have different export capabilities, with L7Ae exporting twice as many transcripts per cell compared to MCP.

Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
References
- ↑ Ljubetič, A., et al. (2017). "Design of coiled-coil protein-origami cages that self-assemble in vitro and in vivo." Nature Biotechnology 35: 1094.
