Difference between revisions of "Part:BBa K2933001"
(→Enzyme activity determination) |
(→Conclusion) |
||
| (254 intermediate revisions by 7 users not shown) | |||
| Line 14: | Line 14: | ||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
| − | ===Functional Parameters=== | + | `2===Functional Parameters=== |
<partinfo>BBa_K2933001 parameters</partinfo> | <partinfo>BBa_K2933001 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| − | NDM-23 is a type of subclass | + | NDM-23 is a type of subclass B1 metallo-beta-lactamases, which is derived from NDM-1 mutation. The beta lactamases of the NDM family can hydrolyze almost all available beta lactam antibiotics (except aztreonam) clinically, including the broad-spectrum antibiotic carbapenems. Because of the extensive substrate profile of this enzyme, the clinical strains carrying it become a great threat to human life and health.<br> |
| − | + | In 2008, Dongeun Yong and his team first discovered NDM-1 in a Klebsiella pneumoniae isolated from a Swedish patient who was hospitalized in New Delhi, India. Since then, under the pressure of selection of antibacterial drugs, NDM has been reported worldwide, and 24 variants have been discovered so far. Studies have shown that blaNDM in CRE strain often coexists with blaOXA, blaSHV, blaVIM and other drug resistance genes, making the carrying strain multi-drug resistant, and even with the mcr-1 gene at the same time, it is pan-resistant, greatly limit the choice and use of clinical antibacterial drugs.<br> | |
| − | + | According to statistics, Enterobacteriaceae producing NDM-type carbapenemase have been used in various Enterobacteriaceae such as Escherichia coli, Klebsiella pneumoniae, Klebsiella oxysporum and Citrobacter freundii. It spreads widely and is widely distributed in many countries around the world such as India, Brazil, the United States, France and China.<br> | |
| − | + | ||
| − | + | ||
| − | ---- | + | |
| + | ===Origin(organism)=== | ||
| + | Klebsiella pneumoniae.<br> | ||
===Molecular cloning=== | ===Molecular cloning=== | ||
| − | + | First, we used the vector pGEX-6p-1 to construct our expression plasmid. And then we converted the plasmid constructed to ''E. coli'' DH5α to expand the plasmid largely.<br> | |
| + | <p style="text-align: center;"> | ||
| + | [[File:NDM-23-PCR.png|500px]]<br> | ||
| + | '''Figure 1.''' Left: The PCR result of NDM-23. Right: The verification results by enzyme digestion.<br> | ||
| + | </p> | ||
| + | After verification, it was determined that the construction is successful. We converted the plasmid to ''E. coli'' BL21(DE3) for expression and purification.<br> | ||
===Expression and purification=== | ===Expression and purification=== | ||
| − | Pre-expression:<br> | + | '''Pre-expression:'''<br> |
The bacteria were cultured in 5mL LB liquid medium with ampicillin(100 μg/mL final concentration) in 37℃ overnight.<br> | The bacteria were cultured in 5mL LB liquid medium with ampicillin(100 μg/mL final concentration) in 37℃ overnight.<br> | ||
| + | '''Massive expressing:'''<br> | ||
| + | After taking samples, we transfered them into 1L LB medium and added antibiotic to 100 μg/mL final concentration. Grow them up in 37°C shaking incubator. Grow until an OD 600 nm of 0.8 to 1.2 (roughly 3-4 hours). Induce the culture to express protein by adding 1 mM IPTG (isopropylthiogalactoside, MW 238 g/mol). Put the liter flasks in 16°C shaking incubator for 16h.<br> | ||
| − | + | '''Affinity Chromatography:'''<br> | |
| − | + | We used the GST Agarose to purify the target protein. The GST Agarose can combine specifically with the GST tag fused with target protein. <br> | |
| + | * First, wash the column with GST-binding buffer for 10 minutes to balance the GST column.<br> | ||
| + | * Second, add the protein solution to the column, let it flow naturally and bind to the column.<br> | ||
| + | * Third, add GST-Washing buffer several times and let it flow. Take 10μl of wash solution and test with Coomassie Brilliant Blue. Stop washing when it doesn’t turn blue.<br> | ||
| + | * Forth, add 400μL Prescission Protease (1mg/mL) to the agarose. Digest for 16 hours in 4℃. | ||
| + | * Fifth, add GST-Elution buffer several times. Check as above. Collect the eluted proteins for further operation.<br> | ||
| + | <p style="text-align: center;"> | ||
| + | [[File:T--TJUSLS China--NDM 23 GST.jpg|400px]]<br> | ||
| − | + | '''Figure 2.''' The result of SDS-page.<br> | |
| − | + | </p> | |
| − | Anion exchange column:<br> | + | '''Anion exchange column:'''<br> |
| − | According to the predicted pI of the protein and the pH of the ion-exchange column buffer, firstly select the appropriate ion exchange column (anion exchange column or cation exchange column). The pH of buffer should deviate from the isoelectric point of the protein. Since the isoelectric point of our protein is | + | According to the predicted pI of the protein and the pH of the ion-exchange column buffer, firstly select the appropriate ion exchange column (anion exchange column or cation exchange column). The pH of buffer should deviate from the isoelectric point of the protein. Since the isoelectric point of our protein is 5.88 in theory, we choose buffer pH of 7.4 and use anion exchange column for purification. |
The protein is concentrated with a 10KD concentration tube, and then the exchange buffer is used to exchange the protein to the ion-exchange liquid A. Finally, it is concentrated to less than 5ml by centrifuging at 4℃ and 3400rpm for 10 minutes in a high-speed centrifuge to remove insoluble substances and bubbles. | The protein is concentrated with a 10KD concentration tube, and then the exchange buffer is used to exchange the protein to the ion-exchange liquid A. Finally, it is concentrated to less than 5ml by centrifuging at 4℃ and 3400rpm for 10 minutes in a high-speed centrifuge to remove insoluble substances and bubbles. | ||
Balance the selected column with liquid A. Through the AKTApure protein purification system, the samples are loaded to the column at a flow rate of 0.5ml/min, and continue washing for 5min. Gradually increase the content of liquid B in the column, change the salt concentration and then change the interaction between the sample and the column, and collect the corresponding eluent according to the position of the peak. Use SDS-PAGE to check the result.<br> | Balance the selected column with liquid A. Through the AKTApure protein purification system, the samples are loaded to the column at a flow rate of 0.5ml/min, and continue washing for 5min. Gradually increase the content of liquid B in the column, change the salt concentration and then change the interaction between the sample and the column, and collect the corresponding eluent according to the position of the peak. Use SDS-PAGE to check the result.<br> | ||
| − | + | <p style="text-align: center;"> | |
| − | Gel filtration chromatography:<br> | + | [[File:T--TJUSLS China--NDM 23 Q.jpg|400px|]]<br> |
| + | '''Figure 3.''' The result of SDS-PAGE of Q column.<br> | ||
| + | </p> | ||
| + | '''Gel filtration chromatography:'''<br> | ||
The collected protein samples are concentrated in a 10 KD concentrating tube at a speed of 3400 rpm and concentrated for a certain time until the sample volume is 500 μl. At the same time, the superdex 200 column is equilibrated with a buffer to balance 1.2 column volumes. The sample is then loaded and 1.5 cylinders are eluted isocratically with buffer. Determine the state of protein aggregation based on the peak position and collect protein samples based on the results of running the gel.<br> | The collected protein samples are concentrated in a 10 KD concentrating tube at a speed of 3400 rpm and concentrated for a certain time until the sample volume is 500 μl. At the same time, the superdex 200 column is equilibrated with a buffer to balance 1.2 column volumes. The sample is then loaded and 1.5 cylinders are eluted isocratically with buffer. Determine the state of protein aggregation based on the peak position and collect protein samples based on the results of running the gel.<br> | ||
| + | |||
<p style="text-align: center;"> | <p style="text-align: center;"> | ||
| − | [[File:T--TJUSLS China--NDM 23 gel jiaotu+fengtu.png]]<br> | + | [[File:T--TJUSLS China--NDM 23 gel jiaotu+fengtu.png|600px|]]<br> |
| − | '''Figure | + | '''Figure 4.''' (a) The result of gel filtration used the superdex75 column with the AKTA system, which shows that the target protein is monomeric. (b) The result of SDS-PAGE. And the target protein is about 28.5kD.<br> |
</p> | </p> | ||
===Enzyme activity determination=== | ===Enzyme activity determination=== | ||
| − | We | + | We used CDC-1, a probe with a similar structure from the beta lactam ring and a luminescent group for enzyme activity measurements. For more information on the substrate CDC-1, please see our project introduction.<br> |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | Buffer: | + | '''Materials:'''<br> |
| − | 100% DMSO | + | General 96-well plates (Black)<br> |
| − | Fluorescent Probe(CDC-1) | + | Infinite M1000 Pro Automatic Microplate Reader<br> |
| − | Target Enzyme(beta-lactamase) | + | Multi-channel adjustable pipette<br> |
| + | Ultrasonic Cleaner<br> | ||
| + | |||
| + | '''Buffer:'''<br> | ||
| + | 100% DMSO<br> | ||
| + | Fluorescent Probe(CDC-1)<br> | ||
| + | Target Enzyme(beta-lactamase)<br> | ||
====Determination of enzyme concentration==== | ====Determination of enzyme concentration==== | ||
| + | <p style="text-align: center;"> | ||
| + | [[File:NDM.jpg|300px|]][[File:NDM TXSX.jpeg|300px|]]<br> | ||
| + | '''Figure 5.''' The concentration of CDC-1 was fixed at 10.5 μM and the enzyme concentration was changed within a certain range, and the fluorescence value was measured with a function of reaction time. Left: First, we selected three gradient concentrations (with large intervals) for pre-experiment, and determined the gradient range of the formal experiment through the experimental results. Right: The appropriate enzyme concentration was selected for determination of the gradient, and the reaction curve of gradual rise was obtained.<br> | ||
| + | </p> | ||
| + | <p style="text-align: center;"> | ||
| + | [[File:T--TJUSLS China--NDM 23 EC80.png|300px]]<br> | ||
| + | '''Figure 6.''' We took the emission fluorescence at 3.02nm as the maximum emission fluorescence, and took the logarithm value of different NDM-23 enzyme concentrations to make the relationship curve between protein concentration and fluorescence emission rate. When the emittance of the system was 80%, the protein concentration was 1.648nM.<br> | ||
| + | </p> | ||
| − | ====Determination of the buffer | + | ====Determination of the buffer condition==== |
| + | <p style="text-align: center;"> | ||
| + | [[File:Ndm zn new.png|600px]]<br> | ||
| + | '''Figure 7.''' Effect of different buffer condition on enzyme activity.<br> | ||
| + | </p> | ||
| + | According to the experimental results, we chose NaCl concentration of 140mM, ZnCl<sub>2 </sub>concentration of 25 μM and pH of 8.0. The effect of DMSO on protein activity can be excluded in the range of 2-10%. (6% in the system)<br> | ||
| − | ==== | + | ====Lineweaver-Burk plot==== |
| + | <p style="text-align: center;"> | ||
| + | [[File:T--TJUSLS China--NDM 23 M.png|300px]]<br> | ||
| + | '''Figure 8.''' The relationship between the substrate concentration and the maximum initial rate was obtained by using the Lineweaver-Burk plot.<br> | ||
| + | </p> | ||
| + | <p style="text-align: center;"> | ||
| + | [[File:T--TJUSLS China--NDM 23 Kcat.png|300px]]<br> | ||
| + | '''Figure 9.''' The relationship between the maximum fluorescence value and substrate concentration. | ||
| + | </p> | ||
| + | Calculate Km, Vm with the Lineweaver-Burk plot, because it fit better. Kcat values were calculated with the results of maximum fluorescence values at different substrate concentrations.<br> | ||
| + | <p style="text-align: center;"> | ||
| + | [[File:T--TJUSLS China--NDM 23 Km Kcat.png|400px]]<br> | ||
| + | '''Figure 10.''' The enzyme kinetic parameter of NDM-23.<br> | ||
| + | </p> | ||
| − | ==== | + | ===Establishment of NDM-23 inhibitor screening system=== |
| + | After the above determination of enzyme activity and the trial of concentration and buffer components, we determined the optimal conditions of NDM-23 enzyme activity and then established the screening system.<br> | ||
| + | <p style="text-align: center;"> | ||
| + | [[File:T--TJUSLS China--NDM 23 screen system.png|300px]]<br> | ||
| + | '''Figure11.''' Protein concentration and optimal buffer components of NDM-23. | ||
| + | </p> | ||
| + | |||
| + | <p style="text-align: center;"> | ||
| + | [[File:T--TJUSLS China--NDM 23 screen system1u.png|400px]]<br> | ||
| + | '''Figure12.''' The inhibitor screening system of NDM-23. | ||
| + | |||
| + | ===Effective inhibitors in vitro we found=== | ||
| + | Above, we have established the NDM-23 high-throughput screening system, and then we used the microplate reader to conduct high-throughput screening to screen out '''20''' inhibitors with significant inhibitory effect on NDM-23 from the drug library containing over '''4000''' small molecules.<br> | ||
| + | [[File:T--TJUSLS China--NDM 23 inhibitor1.png|400px]] [[File:T--TJUSLS China--NDM 23 inhibitor2.png|400px]]<br> | ||
| + | |||
| + | [[File:T--TJUSLS China--NDM 23 inhibitor3.png|400px]] [[File:T--TJUSLS China--NDM 23 inhibitor4.png|400px]]<br> | ||
| + | |||
| + | [[File:T--TJUSLS China--NDM 23 inhibitor5.png|400px]] [[File:T--TJUSLS China--NDM 23 inhibitor6.png|400px]]<br> | ||
| + | |||
| + | [[File:T--TJUSLS China--NDM 23 inhibitor7.png|400px]] [[File:T--TJUSLS China--NDM 23 inhibitor8.png|400px]]<br> | ||
| + | |||
| + | [[File:T--TJUSLS China--NDM 23 inhibitor9.png|400px]] [[File:T--TJUSLS China--NDM 23 inhibitor10.png|400px]]<br> | ||
| + | |||
| + | [[File:T--TJUSLS China--NDM 23 inhibitor13.png|400px]] [[File:T--TJUSLS China--NDM 23 inhibitor12.png|400px]]<br> | ||
| + | |||
| + | [[File:T--TJUSLS China--NDM 23 inhibitor15.png|400px]] [[File:T--TJUSLS China--NDM 23 inhibitor16.png|400px]]<br> | ||
| + | |||
| + | [[File:T--TJUSLS China--NDM 23 inhibitor17.png|400px]] [[File:T--TJUSLS China--NDM 23 inhibitor18.png|400px]]<br> | ||
| + | |||
| + | [[File:T--TJUSLS China--NDM 23 inhibitor19.png|400px]] [[File:T--TJUSLS China--NDM 23 inhibitor20.png|400px]]<br> | ||
| + | [[File:T--TJUSLS China--NDM 23 inhibitor14.png|400px]] [[File:Silver NDM YIZHIQVXIAN.png|400px|]]<br> | ||
| + | |||
| + | ===Extracorporeal IC<sub>50</sub> and inhibitory mechanism of inhibitors=== | ||
| + | We tested the IC<sub>50</sub> of eight inhibitors and the inhibitory mechanism of 6 inhibitors.<br> | ||
| + | </p> | ||
| + | <p style="text-align: center;"> | ||
| + | [[File:TA NDM IC50.jpeg|300px]][[File:TA reversible inhibition.jpeg|300px]][[File:TA competitive inhibition.jpeg|300px]]<br> | ||
| + | '''Figure 13.''' IC<sub>50</sub> and inhibitory mechanism of Tannic acid for NDM-23. Its inhibition type is competitive inhibition of reversible inhibition.<br> | ||
| + | </p> | ||
| + | <p style="text-align: center;"> | ||
| + | [[File:EGCG NDM IC50.jpeg|300px|]][[File:EGCG Reversible Inhibition.jpeg|300px|]][[File:EGCG competitive.jpeg|300px|]]<br> | ||
| + | '''Figure 14.''' IC<sub>50</sub> and inhibitory mechanism of (-)-Epigallocatechin gallate(EGCG) for NDM-23.Its inhibition type is competitive inhibition of reversible inhibition.<br> | ||
| + | </p> | ||
| + | <p style="text-align: center;"> | ||
| + | [[File:Corilagin NDM IC50.jpeg|300px|]][[File:KLLJ reversible.jpeg|300px|]][[File:KLLJ uncompetitive.jpeg|300px|]]<br> | ||
| + | '''Figure 15.''' IC<sub>50</sub> and inhibitory mechanism of Corilagin for NDM-23.Its inhibition type is uncompetitive inhibition of reversible inhibition.<br> | ||
| + | </p> | ||
| + | <p style="text-align: center;"> | ||
| + | [[File:Hamdy IC50NDM.jpeg|300px|]][[File:Silver sulfadiazine NDM keni.jpeg|300px|]][[File:Silver sulfadiazine NDM feijingzheng.jpeg|300px|]]<br> | ||
| + | '''Figure 16.''' IC<sub>50</sub> and inhibitory mechanism of silver sulfadiazine for NDM-23.Its inhibition type is uncompetitive inhibition of reversible inhibition.<br> | ||
| + | </p> | ||
| + | <p style="text-align: center;"> | ||
| + | [[File:IC50HCL.jpeg|300px|]][[File:HCL keni.jpeg|300px|]][[File:Yansuanf.jpeg|300px|]]<br> | ||
| + | '''Figure 17.''' IC<sub>50</sub> and inhibitory mechanism of Fingolimod hydrochloride for NDM-23.Its inhibition type is competitive inhibition of reversible inhibition.<br> | ||
| + | </p> | ||
| + | <p style="text-align: center;"> | ||
| + | [[File:YXSIC50.jpeg|300px|]][[File:Ginkgoic acid IRRESIBLE.jpeg|300px|]]<br> | ||
| + | '''Figure 18.''' IC<sub>50</sub> and inhibitory mechanism of ginkgoic acid for NDM-23.Its inhibition type is irreversible inhibition.<br> | ||
| + | </p> | ||
| + | <p style="text-align: center;"> | ||
| + | [[File:Etravirine IC50.jpeg|300px|]]<br> | ||
| + | '''Figure 19.''' IC<sub>50</sub> of Etravirine for NDM-23.<br> | ||
| + | </p> | ||
| + | <p style="text-align: center;"> | ||
| + | [[File:Adapalene NDM IC50.jpeg|300px|]]<br> | ||
| + | '''Figure 20.''' IC<sub>50</sub> of Adapalene for NDM-23.<br> | ||
| + | </p> | ||
| + | |||
| + | ===Monitoring in living bacterial cells with antibiotics=== | ||
| + | 1.Experiments proving the accuracy of the system<br> | ||
| + | Firstly, NDM-23 protein solution at 3.02nM was mixed with cefazolin at 250 μM, and the characteristic absorption peak of cefazolin at 273nm obtained by our experiment was continuously measured.<br> | ||
| + | <p style="text-align: center;"> | ||
| + | [[File:Data 1 NMD-23.jpeg|300px|]][[File:NDM time1.png|300px|]]<br> | ||
| + | '''Figure 21.''' The curve of specific absorption peak of antibiotics mixed with NDM-23 was revealed. | ||
| + | </p> | ||
| + | Secondly, after phosphate buffer treatment, E. coli strain transferred to NDM-23 beta-lactamase gene was diluted to OD600=0.15, and the strain was mixed with cefazolin with a final concentration of 250 μM, and the peak value of cefazolin at 273nm was continuously measured. The results are shown below.<br> | ||
| + | <p style="text-align: center;"> | ||
| + | [[File:Data2 NDM-23.jpeg|300px|]][[File:Time ndm 2.png|300px|]]<br> | ||
| + | '''Figure 22.'''The curve of specific absorption peak of antibiotics mixed with E. coli with the NDM23 beta-lactamase gene was revealed.<br> | ||
| + | At the same time, the phosphate buffer used in the preparation of bacterial solution and the E.coli without the transfer of NDM-23 beta-lactamase gene were mixed with cefazolin with a final concentration of 250 μM respectively as blank control and negative control, and the specific absorption peak value of cefazolin at 273nm was continuously determined.<br> | ||
| + | |||
| + | [[File:Data3NDM23.jpeg|300px|]][[File:Time3ndm.png|300px|]]<br> | ||
| + | [[File:Data4NDM.jpeg|300px|]][[File:NDM Time4.png|300px|]]<br> | ||
| + | '''Figure 23.''' The curve of specific absorption peak of antibiotics mixed with the phosphate buffer and the E.coli without the transfer of NDM23 beta-lactamase gene was revealed. The one up is antibiotic combined with phosphate buffer and the one down is antibiotic combined with the E coli without the transfer of NDM-23 beta-lactamase gene.<br> | ||
| + | </p> | ||
| + | 2.Hydrolysis efficiency to different antibiotics<br> | ||
| + | [[File:T--TJUSLS China--NDM 23 different β2.png|300px|center|]]<br> | ||
| + | '''Figure 24.''' The stability of four antibiotics in the presence of the NDM23 ''E. coli'' cells. The hydrolysis of antibiotics was monitored through absorbance changes of the drugs at 273, 300,307 and 360 nm for cefazolin, meropenem and faropenem, and tetracycline,respectively,in the kinetic mode (120min, triplicate scans per 5 min). The starting concentration of antibiotics was 250 μM.<br> | ||
| + | |||
| + | 3.Evaluation of therapeutic effect of NDM-23<br> | ||
| + | After determining the reaction system, we diluted the concentration gradient of several inhibitors and incubated them with the prepared E.coli which has been transferred into NDM23 beta-lactamase gene in dark for 1h, then added cefazolin with a final concentration of 250 μM and continuously measured the light absorption value at 273nm. The results are as follows:<br> | ||
| + | <p style="text-align: center;"> | ||
| + | [[File:NDM23 EDTA.jpeg|300px|]]<br> | ||
| + | '''Figure 25.''' Monitoring in living bacterial cells with antibiotics and EDTA(positive control).<br> | ||
| + | </p> | ||
| + | <p style="text-align: center;"> | ||
| + | [[File:KLLJhuojunjianc.jpeg|300px|]]<br> | ||
| + | '''Figure 26.''' Monitoring in living bacterial cells with antibiotics and Corilagin.<br> | ||
| + | </p> | ||
| + | <p style="text-align: center;"> | ||
| + | [[File:EGCGhuojun.jpeg|300px|]]<br> | ||
| + | '''Figure 27.''' Monitoring in living bacterial cells with antibiotics and EGCG.<br> | ||
| + | </p> | ||
| + | <p style="text-align: center;"> | ||
| + | [[File:Tannic acid huojun.jpeg|300px|]]<br> | ||
| + | '''Figure 28.''' Monitoring in living bacterial cells with antibiotics and Tannic acid.<br> | ||
| + | </p> | ||
| − | === | + | ===Conclusion=== |
| + | In conclusion, NDM-23 protein was successfully expressed in this part. We measured enzyme activity, established the high-throughput screening system, successfully screened some effective inhibitors with CDC-1 probes and then verified some of them with live bacteria to determine the IC<sub>50</sub> of the inhibitors in vivo. We found that the inhibitors can effectively inhibit the activity of the enzyme in vivo and prevent the hydrolysis of cefazolin by the enzyme. We are proud that our results have laid the foundation for further research. | ||
| − | === | + | ===Reference=== |
| + | [1] Van Duin D, Doi Y. The global epidemiology of carbapenemase-producing Enterobacteriaceae [J]. Virulence, 2017,8(4): 460469.<br> | ||
| + | [2] Yong D, Toleman MA, Giske cG, et al. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique geneticstructure in Klebsiella pneumoniae sequence type 14 from India [J]. Antimicrob Agents Chemother, 2009,53(12): 5046-5054.<br> | ||
| + | [3] Wu w. Feng Y, Tang G et al. NDM Metallo-ß-Lactamases and Their Bacterial Producers in Health Care Sttings [J]. Clin Microbiol Rev, 2019,32(2): 0011500118.<br> | ||
| + | [4] Khan AU, Maryam L, Zarilli R. Structure, Genetics and Worldwide Spread of New Delhi Maeallo-beta-lactamase (NDM): a threat to public health [J].BMC Microbiol, 2017,17(1):101-112.<br> | ||
| + | [5] Zheng B, Lv T, Xu H, et al. Discovery and characterisation of an escherichia coli ST206 strain producing NDM-5 and MCR-1 from a patient with acute diarrhoea in China [J]. Int JAntimicrob Agents, 2018,51(2): 273-275.<br> | ||
| + | [6] Li X, Jiang Y, Wu K,et al. Whole-genome sequencing identification of a multidrug-resistan t Salmonella enterica serovar Typhimurium strain carrying blaNDM-5 from Guangdong, China [J]. Infect Genet Evol, 2017,55: 195-198.<br> | ||
| + | [7] Rahman M, Shukla SK, Prasad KN, et al. Prevalence and molecular characterisation of New Delhi metallo-β-lactamases NDM-I, NDM-5, NDM-6 and NDM-7 in multidrug- resistant Enterobacteriaceae from India [J]. Int J Antimierob Agents, 2014,44(1).<br> | ||
| + | [8] Rojas LJ, Hujer AM, Rudin SD, et al. NDM-5 and OXA-181 Beta-Lactamases, a Significant Threat Continues To Spread in the Americas [J]. Antimicrob Agents Chemother,2017,61(7): pii: e00454-17. <br> | ||
| + | [9] Almakki A, Maure A, Pantel A, et al. NDM-5-producing Escherichia coli in an urban river in Montpellier, France [I]. Int J Antimicrob Agents, 2017,50(1): 123-124.<br> | ||
| + | [10] Rozales FP, Magagnin cM, Campos JC, et al. Characterization of Transformants Obtained From NDM-1-Producing Enterobacteriaceae in Brazil [J]. Infect Control Hosp Epidemiol,2017,38(5): 634-636.<br> | ||
| + | [11] Yang B, Feng Y, McNally A, et al. Occurrence of Enterobacter hormaechei carrying blaNDM-1 and blaKPC-2 in China [J]. Diagn Microbiol Infect Dis, 2018.90(2): 139-142.<br> | ||
Latest revision as of 13:12, 21 October 2019
subclass B1 metallo-beta-lactamase NDM-23, codon optimized in E. coli
This part encodes a protein called NDM-23, which is a metallo-beta-lactamase of subclass B1.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Usage and Biology
NDM-23 is a type of subclass B1 metallo-beta-lactamases, which is derived from NDM-1 mutation. The beta lactamases of the NDM family can hydrolyze almost all available beta lactam antibiotics (except aztreonam) clinically, including the broad-spectrum antibiotic carbapenems. Because of the extensive substrate profile of this enzyme, the clinical strains carrying it become a great threat to human life and health.
In 2008, Dongeun Yong and his team first discovered NDM-1 in a Klebsiella pneumoniae isolated from a Swedish patient who was hospitalized in New Delhi, India. Since then, under the pressure of selection of antibacterial drugs, NDM has been reported worldwide, and 24 variants have been discovered so far. Studies have shown that blaNDM in CRE strain often coexists with blaOXA, blaSHV, blaVIM and other drug resistance genes, making the carrying strain multi-drug resistant, and even with the mcr-1 gene at the same time, it is pan-resistant, greatly limit the choice and use of clinical antibacterial drugs.
According to statistics, Enterobacteriaceae producing NDM-type carbapenemase have been used in various Enterobacteriaceae such as Escherichia coli, Klebsiella pneumoniae, Klebsiella oxysporum and Citrobacter freundii. It spreads widely and is widely distributed in many countries around the world such as India, Brazil, the United States, France and China.
Origin(organism)
Klebsiella pneumoniae.
Molecular cloning
First, we used the vector pGEX-6p-1 to construct our expression plasmid. And then we converted the plasmid constructed to E. coli DH5α to expand the plasmid largely.

Figure 1. Left: The PCR result of NDM-23. Right: The verification results by enzyme digestion.
After verification, it was determined that the construction is successful. We converted the plasmid to E. coli BL21(DE3) for expression and purification.
Expression and purification
Pre-expression:
The bacteria were cultured in 5mL LB liquid medium with ampicillin(100 μg/mL final concentration) in 37℃ overnight.
Massive expressing:
After taking samples, we transfered them into 1L LB medium and added antibiotic to 100 μg/mL final concentration. Grow them up in 37°C shaking incubator. Grow until an OD 600 nm of 0.8 to 1.2 (roughly 3-4 hours). Induce the culture to express protein by adding 1 mM IPTG (isopropylthiogalactoside, MW 238 g/mol). Put the liter flasks in 16°C shaking incubator for 16h.
Affinity Chromatography:
We used the GST Agarose to purify the target protein. The GST Agarose can combine specifically with the GST tag fused with target protein.
- First, wash the column with GST-binding buffer for 10 minutes to balance the GST column.
- Second, add the protein solution to the column, let it flow naturally and bind to the column.
- Third, add GST-Washing buffer several times and let it flow. Take 10μl of wash solution and test with Coomassie Brilliant Blue. Stop washing when it doesn’t turn blue.
- Forth, add 400μL Prescission Protease (1mg/mL) to the agarose. Digest for 16 hours in 4℃.
- Fifth, add GST-Elution buffer several times. Check as above. Collect the eluted proteins for further operation.

Figure 2. The result of SDS-page.
Anion exchange column:
According to the predicted pI of the protein and the pH of the ion-exchange column buffer, firstly select the appropriate ion exchange column (anion exchange column or cation exchange column). The pH of buffer should deviate from the isoelectric point of the protein. Since the isoelectric point of our protein is 5.88 in theory, we choose buffer pH of 7.4 and use anion exchange column for purification.
The protein is concentrated with a 10KD concentration tube, and then the exchange buffer is used to exchange the protein to the ion-exchange liquid A. Finally, it is concentrated to less than 5ml by centrifuging at 4℃ and 3400rpm for 10 minutes in a high-speed centrifuge to remove insoluble substances and bubbles.
Balance the selected column with liquid A. Through the AKTApure protein purification system, the samples are loaded to the column at a flow rate of 0.5ml/min, and continue washing for 5min. Gradually increase the content of liquid B in the column, change the salt concentration and then change the interaction between the sample and the column, and collect the corresponding eluent according to the position of the peak. Use SDS-PAGE to check the result.

Figure 3. The result of SDS-PAGE of Q column.
Gel filtration chromatography:
The collected protein samples are concentrated in a 10 KD concentrating tube at a speed of 3400 rpm and concentrated for a certain time until the sample volume is 500 μl. At the same time, the superdex 200 column is equilibrated with a buffer to balance 1.2 column volumes. The sample is then loaded and 1.5 cylinders are eluted isocratically with buffer. Determine the state of protein aggregation based on the peak position and collect protein samples based on the results of running the gel.

Figure 4. (a) The result of gel filtration used the superdex75 column with the AKTA system, which shows that the target protein is monomeric. (b) The result of SDS-PAGE. And the target protein is about 28.5kD.
Enzyme activity determination
We used CDC-1, a probe with a similar structure from the beta lactam ring and a luminescent group for enzyme activity measurements. For more information on the substrate CDC-1, please see our project introduction.
Materials:
General 96-well plates (Black)
Infinite M1000 Pro Automatic Microplate Reader
Multi-channel adjustable pipette
Ultrasonic Cleaner
Buffer:
100% DMSO
Fluorescent Probe(CDC-1)
Target Enzyme(beta-lactamase)
Determination of enzyme concentration


Figure 5. The concentration of CDC-1 was fixed at 10.5 μM and the enzyme concentration was changed within a certain range, and the fluorescence value was measured with a function of reaction time. Left: First, we selected three gradient concentrations (with large intervals) for pre-experiment, and determined the gradient range of the formal experiment through the experimental results. Right: The appropriate enzyme concentration was selected for determination of the gradient, and the reaction curve of gradual rise was obtained.

Figure 6. We took the emission fluorescence at 3.02nm as the maximum emission fluorescence, and took the logarithm value of different NDM-23 enzyme concentrations to make the relationship curve between protein concentration and fluorescence emission rate. When the emittance of the system was 80%, the protein concentration was 1.648nM.
Determination of the buffer condition

Figure 7. Effect of different buffer condition on enzyme activity.
According to the experimental results, we chose NaCl concentration of 140mM, ZnCl2 concentration of 25 μM and pH of 8.0. The effect of DMSO on protein activity can be excluded in the range of 2-10%. (6% in the system)
Lineweaver-Burk plot

Figure 8. The relationship between the substrate concentration and the maximum initial rate was obtained by using the Lineweaver-Burk plot.
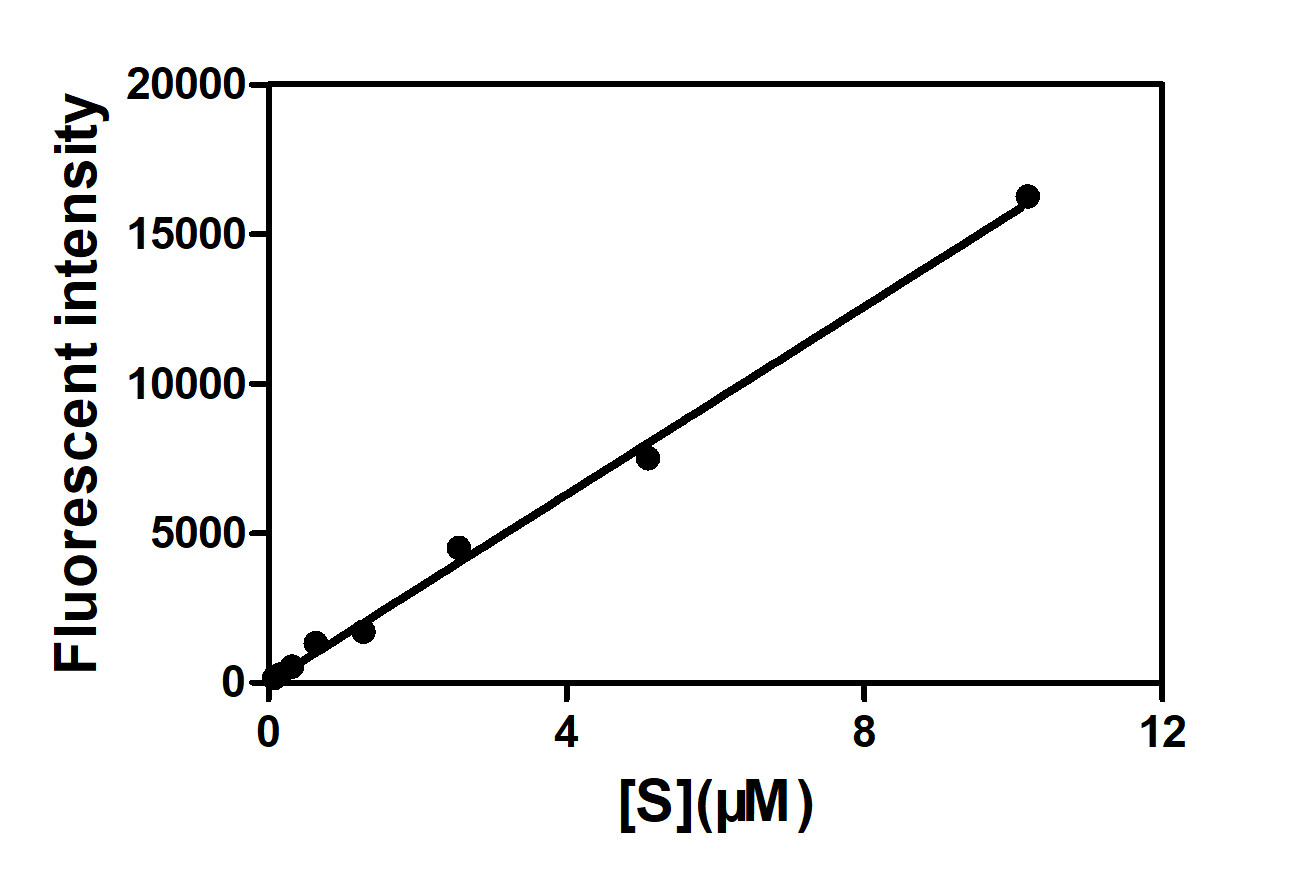
Figure 9. The relationship between the maximum fluorescence value and substrate concentration.
Calculate Km, Vm with the Lineweaver-Burk plot, because it fit better. Kcat values were calculated with the results of maximum fluorescence values at different substrate concentrations.

Figure 10. The enzyme kinetic parameter of NDM-23.
Establishment of NDM-23 inhibitor screening system
After the above determination of enzyme activity and the trial of concentration and buffer components, we determined the optimal conditions of NDM-23 enzyme activity and then established the screening system.

Figure11. Protein concentration and optimal buffer components of NDM-23.

Figure12. The inhibitor screening system of NDM-23.
Effective inhibitors in vitro we found
Above, we have established the NDM-23 high-throughput screening system, and then we used the microplate reader to conduct high-throughput screening to screen out 20 inhibitors with significant inhibitory effect on NDM-23 from the drug library containing over 4000 small molecules.
Extracorporeal IC50 and inhibitory mechanism of inhibitors
We tested the IC50 of eight inhibitors and the inhibitory mechanism of 6 inhibitors.



Figure 13. IC50 and inhibitory mechanism of Tannic acid for NDM-23. Its inhibition type is competitive inhibition of reversible inhibition.



Figure 14. IC50 and inhibitory mechanism of (-)-Epigallocatechin gallate(EGCG) for NDM-23.Its inhibition type is competitive inhibition of reversible inhibition.



Figure 15. IC50 and inhibitory mechanism of Corilagin for NDM-23.Its inhibition type is uncompetitive inhibition of reversible inhibition.



Figure 16. IC50 and inhibitory mechanism of silver sulfadiazine for NDM-23.Its inhibition type is uncompetitive inhibition of reversible inhibition.



Figure 17. IC50 and inhibitory mechanism of Fingolimod hydrochloride for NDM-23.Its inhibition type is competitive inhibition of reversible inhibition.


Figure 18. IC50 and inhibitory mechanism of ginkgoic acid for NDM-23.Its inhibition type is irreversible inhibition.

Figure 19. IC50 of Etravirine for NDM-23.

Figure 20. IC50 of Adapalene for NDM-23.
Monitoring in living bacterial cells with antibiotics
1.Experiments proving the accuracy of the system
Firstly, NDM-23 protein solution at 3.02nM was mixed with cefazolin at 250 μM, and the characteristic absorption peak of cefazolin at 273nm obtained by our experiment was continuously measured.


Figure 21. The curve of specific absorption peak of antibiotics mixed with NDM-23 was revealed.
Secondly, after phosphate buffer treatment, E. coli strain transferred to NDM-23 beta-lactamase gene was diluted to OD600=0.15, and the strain was mixed with cefazolin with a final concentration of 250 μM, and the peak value of cefazolin at 273nm was continuously measured. The results are shown below.


Figure 22.The curve of specific absorption peak of antibiotics mixed with E. coli with the NDM23 beta-lactamase gene was revealed.
At the same time, the phosphate buffer used in the preparation of bacterial solution and the E.coli without the transfer of NDM-23 beta-lactamase gene were mixed with cefazolin with a final concentration of 250 μM respectively as blank control and negative control, and the specific absorption peak value of cefazolin at 273nm was continuously determined.




Figure 23. The curve of specific absorption peak of antibiotics mixed with the phosphate buffer and the E.coli without the transfer of NDM23 beta-lactamase gene was revealed. The one up is antibiotic combined with phosphate buffer and the one down is antibiotic combined with the E coli without the transfer of NDM-23 beta-lactamase gene.
2.Hydrolysis efficiency to different antibiotics
Figure 24. The stability of four antibiotics in the presence of the NDM23 E. coli cells. The hydrolysis of antibiotics was monitored through absorbance changes of the drugs at 273, 300,307 and 360 nm for cefazolin, meropenem and faropenem, and tetracycline,respectively,in the kinetic mode (120min, triplicate scans per 5 min). The starting concentration of antibiotics was 250 μM.
3.Evaluation of therapeutic effect of NDM-23
After determining the reaction system, we diluted the concentration gradient of several inhibitors and incubated them with the prepared E.coli which has been transferred into NDM23 beta-lactamase gene in dark for 1h, then added cefazolin with a final concentration of 250 μM and continuously measured the light absorption value at 273nm. The results are as follows:

Figure 25. Monitoring in living bacterial cells with antibiotics and EDTA(positive control).

Figure 26. Monitoring in living bacterial cells with antibiotics and Corilagin.

Figure 27. Monitoring in living bacterial cells with antibiotics and EGCG.

Figure 28. Monitoring in living bacterial cells with antibiotics and Tannic acid.
Conclusion
In conclusion, NDM-23 protein was successfully expressed in this part. We measured enzyme activity, established the high-throughput screening system, successfully screened some effective inhibitors with CDC-1 probes and then verified some of them with live bacteria to determine the IC50 of the inhibitors in vivo. We found that the inhibitors can effectively inhibit the activity of the enzyme in vivo and prevent the hydrolysis of cefazolin by the enzyme. We are proud that our results have laid the foundation for further research.
Reference
[1] Van Duin D, Doi Y. The global epidemiology of carbapenemase-producing Enterobacteriaceae [J]. Virulence, 2017,8(4): 460469.
[2] Yong D, Toleman MA, Giske cG, et al. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique geneticstructure in Klebsiella pneumoniae sequence type 14 from India [J]. Antimicrob Agents Chemother, 2009,53(12): 5046-5054.
[3] Wu w. Feng Y, Tang G et al. NDM Metallo-ß-Lactamases and Their Bacterial Producers in Health Care Sttings [J]. Clin Microbiol Rev, 2019,32(2): 0011500118.
[4] Khan AU, Maryam L, Zarilli R. Structure, Genetics and Worldwide Spread of New Delhi Maeallo-beta-lactamase (NDM): a threat to public health [J].BMC Microbiol, 2017,17(1):101-112.
[5] Zheng B, Lv T, Xu H, et al. Discovery and characterisation of an escherichia coli ST206 strain producing NDM-5 and MCR-1 from a patient with acute diarrhoea in China [J]. Int JAntimicrob Agents, 2018,51(2): 273-275.
[6] Li X, Jiang Y, Wu K,et al. Whole-genome sequencing identification of a multidrug-resistan t Salmonella enterica serovar Typhimurium strain carrying blaNDM-5 from Guangdong, China [J]. Infect Genet Evol, 2017,55: 195-198.
[7] Rahman M, Shukla SK, Prasad KN, et al. Prevalence and molecular characterisation of New Delhi metallo-β-lactamases NDM-I, NDM-5, NDM-6 and NDM-7 in multidrug- resistant Enterobacteriaceae from India [J]. Int J Antimierob Agents, 2014,44(1).
[8] Rojas LJ, Hujer AM, Rudin SD, et al. NDM-5 and OXA-181 Beta-Lactamases, a Significant Threat Continues To Spread in the Americas [J]. Antimicrob Agents Chemother,2017,61(7): pii: e00454-17.
[9] Almakki A, Maure A, Pantel A, et al. NDM-5-producing Escherichia coli in an urban river in Montpellier, France [I]. Int J Antimicrob Agents, 2017,50(1): 123-124.
[10] Rozales FP, Magagnin cM, Campos JC, et al. Characterization of Transformants Obtained From NDM-1-Producing Enterobacteriaceae in Brazil [J]. Infect Control Hosp Epidemiol,2017,38(5): 634-636.
[11] Yang B, Feng Y, McNally A, et al. Occurrence of Enterobacter hormaechei carrying blaNDM-1 and blaKPC-2 in China [J]. Diagn Microbiol Infect Dis, 2018.90(2): 139-142.





















