Difference between revisions of "Part:BBa B0014"
SunJi UCAS (Talk | contribs) |
|||
| (2 intermediate revisions by 2 users not shown) | |||
| Line 30: | Line 30: | ||
The 2018 Hawaii iGEM team evaluated the 40 most frequently used BioBricks and ran them through an internal priming screening process that we developed using the BLAST program tool. Out of the 40 BioBricks we evaluated, 10 of them showed possible internal priming of either the VF2 or VR primers and sometime even both. The data set has a range of sequence lengths from as small as 12 bases to as large as 1,210 bases. We experienced the issue of possible internal priming during the sequence verification process of our own BBa_K2574001 BioBrick and in the cloning process to express the part as a fusion protein. BBa_K2574001 is a composite part containing a VLP forming Gag protein sequence attached to a frequently used RFP part (BBa_E1010). We conducted a PCR amplification of the Gag-RFP insert using the VF2 and VR primers on the ligation product (pSB1C3 ligated to the Gag + RFP). This amplicon would serve as template for another PCR where we would add the NcoI and BamHI restriction enzyme sites through new primers for ligation into pET14b and subsequent induced expression. Despite gel confirming a rather large, approximately 2.1 kb insert band, our sequencing results with the VR primer and BamHI RFP reverse primer gave mixed results. Both should have displayed the end of the RFP, but the VR primer revealed the end of the Gag. Analysis of the VR primer on the Gag-RFP sequence revealed several sites where the VR primer could have annealed with ~9 - 12 bp of complementarity. Internal priming of forward and reverse primers can be detrimental to an iGEM project because you can never be sure if the desired construct was correctly inserted into the BioBrick plasmid without a successful sequence verification. | The 2018 Hawaii iGEM team evaluated the 40 most frequently used BioBricks and ran them through an internal priming screening process that we developed using the BLAST program tool. Out of the 40 BioBricks we evaluated, 10 of them showed possible internal priming of either the VF2 or VR primers and sometime even both. The data set has a range of sequence lengths from as small as 12 bases to as large as 1,210 bases. We experienced the issue of possible internal priming during the sequence verification process of our own BBa_K2574001 BioBrick and in the cloning process to express the part as a fusion protein. BBa_K2574001 is a composite part containing a VLP forming Gag protein sequence attached to a frequently used RFP part (BBa_E1010). We conducted a PCR amplification of the Gag-RFP insert using the VF2 and VR primers on the ligation product (pSB1C3 ligated to the Gag + RFP). This amplicon would serve as template for another PCR where we would add the NcoI and BamHI restriction enzyme sites through new primers for ligation into pET14b and subsequent induced expression. Despite gel confirming a rather large, approximately 2.1 kb insert band, our sequencing results with the VR primer and BamHI RFP reverse primer gave mixed results. Both should have displayed the end of the RFP, but the VR primer revealed the end of the Gag. Analysis of the VR primer on the Gag-RFP sequence revealed several sites where the VR primer could have annealed with ~9 - 12 bp of complementarity. Internal priming of forward and reverse primers can be detrimental to an iGEM project because you can never be sure if the desired construct was correctly inserted into the BioBrick plasmid without a successful sequence verification. | ||
| + | ==SUSTech_Shenzhen 2021's Characterisation== | ||
| + | |||
| + | We successfully verified the availability of this bidirectional terminator in <i>E. coli</i> Acella. In our experiment, both PhlF and GFP were expressed with the assistance of this terminator in <i>E. coli</i> Acella chassis (figure 2, figure 3). And the correct size of PhlF from western blot indicates an effective termination (figure. 3). | ||
| + | |||
| + | |||
| + | <div align="center" class="img-margin">https://2021.igem.org/File:T--SUSTech_Shenzhen--con_cir.png<p style="font-size: 15px;">Figure 1 PhlF + GFP + PphlF sequence design.</p> | ||
| + | </div> | ||
| + | |||
| + | <div align="center" class="img-margin">https://2021.igem.org/File:T--SUSTech_Shenzhen--part-101wb.jpeg<p style="font-size: 15px;">Fig 2 Western blot of PhlF (E is experiment; C is control; S is supernatant; P is precipitate; PC is positive control). </p> | ||
| + | </div> | ||
| + | |||
| + | <div align="center" class="img-margin">https://2021.igem.org/File:T--SUSTech_Shenzhen--part-cont_fl.png<p style="font-size: 15px;">Figure. 3 Fluorescence test of GFP with different concentrations of IPTG and same concentration of AHL. </p> | ||
| + | </div> | ||
Latest revision as of 09:43, 7 October 2022
|
double terminator (B0012-B0011)
|
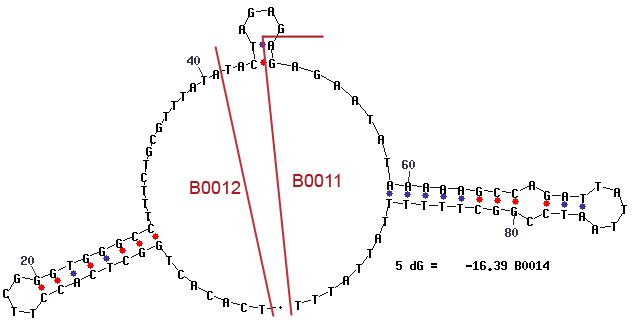
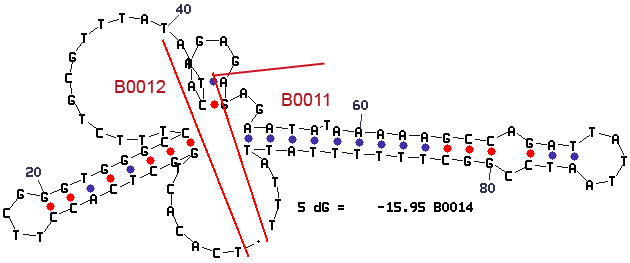
Secondary Structure
The mfold results are annotated with the location of the subparts BBa_B0012 and BBa_B0011.
Measurement
- [http://openwetware.org/wiki/Cconboy:Terminator_Characterization/Results How these parts were measured]
>Internal Priming Screening Characterization of BBa_B0014: Has no possible internal priming sites between this BioBrick part and the VF2 or the VR primer.
The 2018 Hawaii iGEM team evaluated the 40 most frequently used BioBricks and ran them through an internal priming screening process that we developed using the BLAST program tool. Out of the 40 BioBricks we evaluated, 10 of them showed possible internal priming of either the VF2 or VR primers and sometime even both. The data set has a range of sequence lengths from as small as 12 bases to as large as 1,210 bases. We experienced the issue of possible internal priming during the sequence verification process of our own BBa_K2574001 BioBrick and in the cloning process to express the part as a fusion protein. BBa_K2574001 is a composite part containing a VLP forming Gag protein sequence attached to a frequently used RFP part (BBa_E1010). We conducted a PCR amplification of the Gag-RFP insert using the VF2 and VR primers on the ligation product (pSB1C3 ligated to the Gag + RFP). This amplicon would serve as template for another PCR where we would add the NcoI and BamHI restriction enzyme sites through new primers for ligation into pET14b and subsequent induced expression. Despite gel confirming a rather large, approximately 2.1 kb insert band, our sequencing results with the VR primer and BamHI RFP reverse primer gave mixed results. Both should have displayed the end of the RFP, but the VR primer revealed the end of the Gag. Analysis of the VR primer on the Gag-RFP sequence revealed several sites where the VR primer could have annealed with ~9 - 12 bp of complementarity. Internal priming of forward and reverse primers can be detrimental to an iGEM project because you can never be sure if the desired construct was correctly inserted into the BioBrick plasmid without a successful sequence verification.
SUSTech_Shenzhen 2021's Characterisation
We successfully verified the availability of this bidirectional terminator in E. coli Acella. In our experiment, both PhlF and GFP were expressed with the assistance of this terminator in E. coli Acella chassis (figure 2, figure 3). And the correct size of PhlF from western blot indicates an effective termination (figure. 3).
Figure 1 PhlF + GFP + PphlF sequence design.
Fig 2 Western blot of PhlF (E is experiment; C is control; S is supernatant; P is precipitate; PC is positive control).
Figure. 3 Fluorescence test of GFP with different concentrations of IPTG and same concentration of AHL.