Difference between revisions of "Part:BBa K2549006"
(→We prefer mN1c over mN1ce) |
|||
| (2 intermediate revisions by 2 users not shown) | |||
| Line 8: | Line 8: | ||
<partinfo>BBa_K2549006 SequenceAndFeatures</partinfo> | <partinfo>BBa_K2549006 SequenceAndFeatures</partinfo> | ||
| + | ==Characterization By Mutational Landscape by AFCM-Egypt 2023 team== | ||
| + | In order to optimize the function of our parts, we've used the concept of Directed Evolution through applying different mutations and measuring the effects of these mutations on their evolutionary epistatic fitness. As displayed in the chart below, the mutation (V67E) shows the highest epistatic fitness, while the lowest score was associated with the mutation (S312R). | ||
| + | <html><div align="center"style="border:solid #17252A; width:80%;float:center;"><img style=" max-width:850px; | ||
| + | width:100%; | ||
| + | height:auto; | ||
| + | position: relative; | ||
| + | top: 50%; | ||
| + | left: 50%; | ||
| + | transform: translate( -50%); | ||
| + | padding-bottom:25px; | ||
| + | padding-top:25px; | ||
| + | "src="https://static.igem.wiki/teams/4586/wiki/parts-de/mouse-notch2.png"> | ||
| + | <p class=MsoNormal align=center style='text-align:left;border:none;width:98% ;justify-content:center;'><span | ||
| + | lang=EN style='font-size:11.0pt;line-height:115%'>Figure . An illustration of the effects of different mutations on the Epistatic Fitness of Mouth notch. | ||
| + | </span></p></div></html> | ||
| + | ==charactrization of mathematical modeling by AFCM-Egypt 2023 team== | ||
| + | This model is based on an assumption that the external domain activation will lead to transmembrane domain activation of our Syn-Notch receptor which starts from cell to cell interaction till activation of the internal domain ZF21.16VP64. | ||
| + | <html><div align="center"style="border:solid #17252A; width:80%;float:center;"><img style=" max-width:850px; | ||
| + | width:100%; | ||
| + | height:auto; | ||
| + | position: relative; | ||
| + | top: 50%; | ||
| + | left: 50%; | ||
| + | transform: translate( -50%); | ||
| + | padding-bottom:25px; | ||
| + | padding-top:25px; | ||
| + | "src="https://static.igem.wiki/teams/4586/wiki/modeling/pasted-image-1.png"> | ||
| + | <p class=MsoNormal align=center style='text-align:left;border:none;width:98% ;justify-content:center;'><span | ||
| + | lang=EN style='font-size:11.0pt;line-height:115%'>As the binding state occurs, It is evident that the signal is transmitted through the transmembrane domain(mouth notch core) that is resulted from cell to cell interaction.. | ||
| + | </span></p></div></html> | ||
| + | ==Experimental Characterization by AFCM-Egypt== | ||
| + | In order to amplify this DNA part, we used PCR amplification to reach the desired concentration to complete our experiments using specific forward and reverse primers, running the parts on gel electrophoresis as this part presents in lane (P1) including CD8 alpha-his tag-mouse notch core-ZF21.16\VP64, and then measuring the specific concentration of the running part using Real-Time PCR as shown in the following figure. | ||
| + | <html><div align="center"style="border:solid #17252A; width:80%;float:center;"><img style=" max-width:850px; | ||
| + | width:100%; | ||
| + | height:auto; | ||
| + | position: relative; | ||
| + | top: 50%; | ||
| + | left: 50%; | ||
| + | transform: translate( -50%); | ||
| + | padding-bottom:25px; | ||
| + | padding-top:25px; | ||
| + | "src="https://static.igem.wiki/teams/4586/wiki/parts-experiments/pcr-ampli.png"> | ||
| + | <p class=MsoNormal align=center style='text-align:left;border:none;width:98% ;justify-content:center;'><span | ||
| + | lang=EN style='font-size:11.0pt;line-height:115%'> | ||
| + | |||
| + | </span></p></div></html> | ||
| + | <br><br><br><br> | ||
| + | We performed the double digestion method for this part in the prefix and suffix with its specific restriction enzyme and applied this part to gel electrophoresis as shown in the following figure in lane (P1). | ||
| + | <html><div align="center"style="border:solid #17252A; width:80%;float:center;"><img style=" max-width:850px; | ||
| + | width:100%; | ||
| + | height:auto; | ||
| + | position: relative; | ||
| + | top: 50%; | ||
| + | left: 50%; | ||
| + | transform: translate( -50%); | ||
| + | padding-bottom:25px; | ||
| + | padding-top:25px; | ||
| + | "src="https://static.igem.wiki/teams/4586/wiki/parts-experiments/digestion-2.png"> | ||
| + | <p class=MsoNormal align=center style='text-align:left;border:none;width:98% ;justify-content:center;'><span | ||
| + | lang=EN style='font-size:11.0pt;line-height:115%'> | ||
| + | |||
| + | </span></p></div></html> | ||
<!-- Add more about the biology of this part here --> | <!-- Add more about the biology of this part here --> | ||
===Biology=== | ===Biology=== | ||
===== We prefer mN1c over mN1ce ===== | ===== We prefer mN1c over mN1ce ===== | ||
| − | [[File:MouseN1ce1c.png|none|333px|thumb|'''Flow cytometry results of the original | + | [[File:MouseN1ce1c.png|none|333px|thumb|'''Flow cytometry results of the original published SynNotch.''' surEGFP is the surface expressed EGFP, which was used as the antigen. Without antigen (+Mock), the EGFP (Y axis, driven by tTAA released after SynNotch activation) was low, and it went "high" after adding the antigen. More details please visit http://2018.igem.org/Team:Fudan/Results and http://2018.igem.org/Team:Fudan/Optimization .]] |
| − | + | [[Part:BBa_K2549006]] is exactly as the published version, and the only difference was we transiently transfected the cells while the authors created stable cells and picked a cell clone for further experiments. Our results show two things: (1) SynNotch background activation is very high, which is why we put a lot effect in http://2018.igem.org/Team:Fudan/Optimization ; (2) mN1c might be better than mN1ce, not only shorter but a little higher signal-noise-ratio. | |
=====Significance of Notch signaling ===== | =====Significance of Notch signaling ===== | ||
Latest revision as of 20:40, 11 October 2023
mouse Notch1 core
This part is the mouse Notch1 minimal regulatory region[1]. It was used as the transmembrane core domain of the SynNotch. However, original reported design has a high background activation in transfected 293T cells. We have put a lot of effect in optimizing this critical transmembrane domain for Notch receptor.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 592
Characterization By Mutational Landscape by AFCM-Egypt 2023 team
In order to optimize the function of our parts, we've used the concept of Directed Evolution through applying different mutations and measuring the effects of these mutations on their evolutionary epistatic fitness. As displayed in the chart below, the mutation (V67E) shows the highest epistatic fitness, while the lowest score was associated with the mutation (S312R).

Figure . An illustration of the effects of different mutations on the Epistatic Fitness of Mouth notch.
charactrization of mathematical modeling by AFCM-Egypt 2023 team
This model is based on an assumption that the external domain activation will lead to transmembrane domain activation of our Syn-Notch receptor which starts from cell to cell interaction till activation of the internal domain ZF21.16VP64.

As the binding state occurs, It is evident that the signal is transmitted through the transmembrane domain(mouth notch core) that is resulted from cell to cell interaction..
Experimental Characterization by AFCM-Egypt
In order to amplify this DNA part, we used PCR amplification to reach the desired concentration to complete our experiments using specific forward and reverse primers, running the parts on gel electrophoresis as this part presents in lane (P1) including CD8 alpha-his tag-mouse notch core-ZF21.16\VP64, and then measuring the specific concentration of the running part using Real-Time PCR as shown in the following figure.

We performed the double digestion method for this part in the prefix and suffix with its specific restriction enzyme and applied this part to gel electrophoresis as shown in the following figure in lane (P1).

Biology
We prefer mN1c over mN1ce
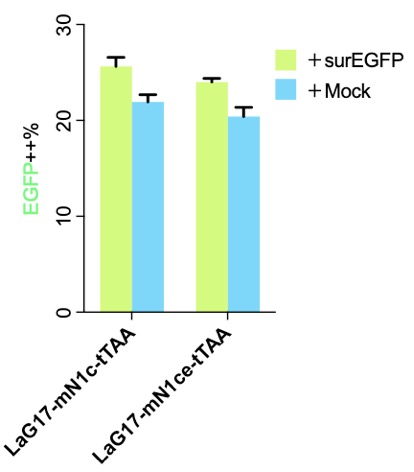
Part:BBa_K2549006 is exactly as the published version, and the only difference was we transiently transfected the cells while the authors created stable cells and picked a cell clone for further experiments. Our results show two things: (1) SynNotch background activation is very high, which is why we put a lot effect in http://2018.igem.org/Team:Fudan/Optimization ; (2) mN1c might be better than mN1ce, not only shorter but a little higher signal-noise-ratio.
Significance of Notch signaling
The Notch signaling pathway is a highly conserved cell signaling system present in most multicellular organisms. Mammals possess four different notch receptors, referred to as NOTCH1 to NOTCH4. The notch receptor is a single-pass transmembrane receptor protein[2].
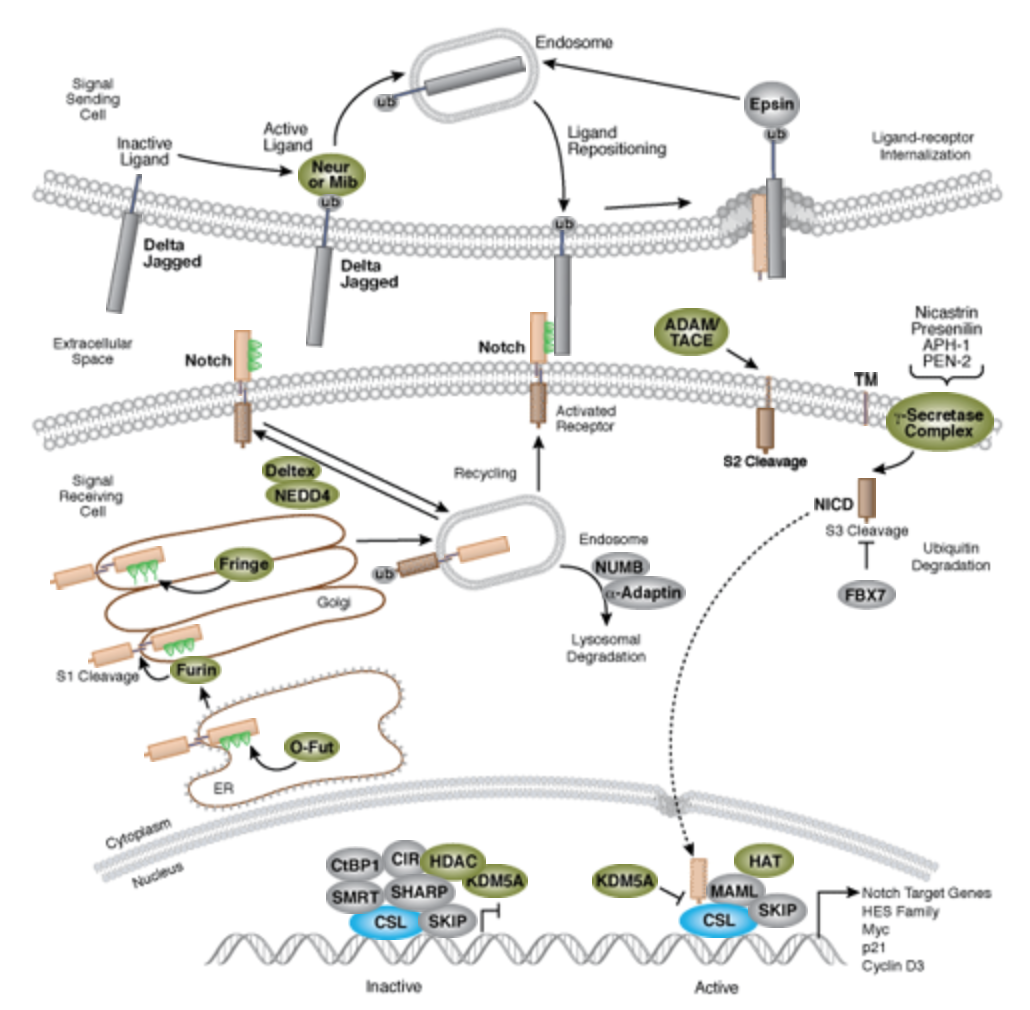
Dr. Hans Widlund (Brigham and Women’s Hospital, Harvard Medical School) once contributed a list (shown below) of original articles worth reading. There are significant amount of Notch research articles and reviews in PubMed[4] as well.
- Ables JL, Breunig JJ, Eisch AJ, Rakic P (2011) Not(ch) just development: Notch signalling in the adult brain. Nat. Rev. Neurosci. 12(5), 269–83.
- Andersson ER, Lendahl U (2014) Therapeutic modulation of Notch signalling--are we there yet? Nat Rev Drug Discov 13(5), 357–78.
- Aster JC, Blacklow SC, Pear WS (2011) Notch signalling in T-cell lymphoblastic leukaemia/lymphoma and other haematological malignancies. J. Pathol. 223(2), 262–73.
- Bai G, Pfaff SL (2011) Protease regulation: the Yin and Yang of neural development and disease. Neuron 72(1), 9–21.
- de la Pompa JL, Epstein JA (2012) Coordinating tissue interactions: Notch signaling in cardiac development and disease. Dev. Cell 22(2), 244–54.
- Ntziachristos P, Lim JS, Sage J, Aifantis I (2014) From fly wings to targeted cancer therapies: a centennial for notch signaling. Cancer Cell 25(3), 318–34.
- Ranganathan P, Weaver KL, Capobianco AJ (2011) Notch signalling in solid tumours: a little bit of everything but not all the time. Nat. Rev. Cancer 11(5), 338–51.
- Weinmaster G, Fischer JA (2011) Notch ligand ubiquitylation: what is it good for? Dev. Cell 21(1), 134–44.
- Yuan JS, Kousis PC, Suliman S, Visan I, Guidos CJ (2010) Functions of notch signaling in the immune system: consensus and controversies. Annu. Rev. Immunol. 28, 343–65.
synNotch with α-CD19 against CD19 antigen works extremely well in Morsut L et al 2016
Please refer the original article for more details.
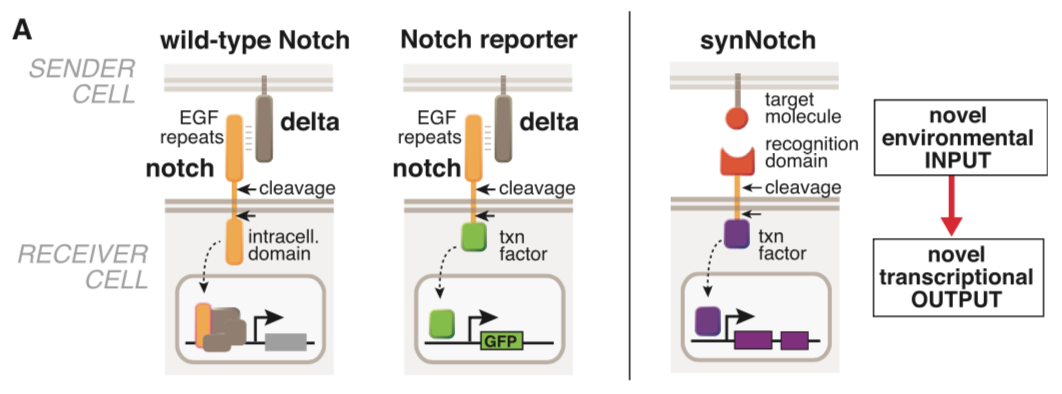
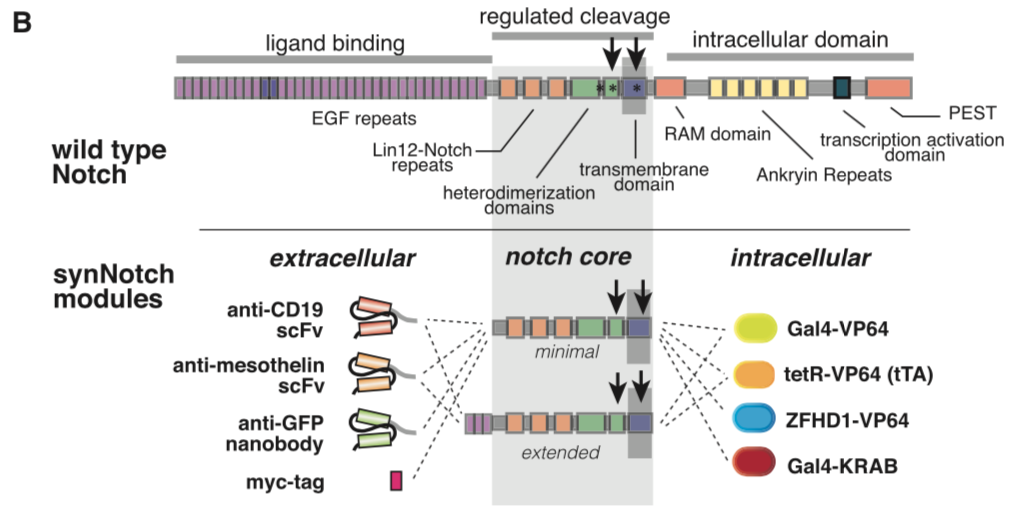





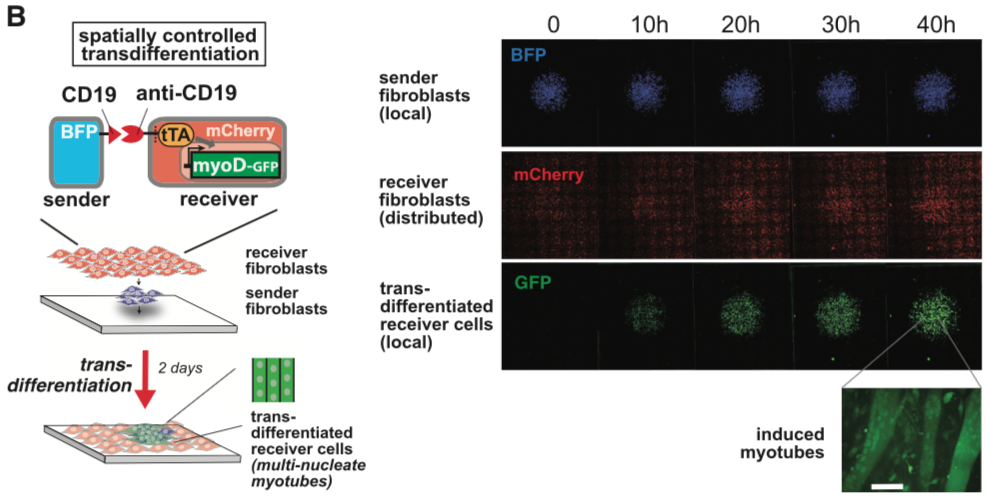

Note: Part:BBa_K2549005 is used in the study described above (Morsut L et al). Part:BBa_K2549007 and Part:BBa_K2549008 have the same Biology section. Our experiments suggested mouse Notch 1 core without extended 3 EGF repeats works better and our receptor optimization all uses Part:BBa_K2549006.
References
- ↑ Engineering Customized Cell Sensing and Response Behaviors Using Synthetic Notch Receptors. Morsut L, Roybal KT, Xiong X, ..., Thomson M, Lim WA. Cell, 2016 Feb;164(4):780-91 PMID: 26830878; DOI: 10.1016/j.cell.2016.01.012
- ↑ https://en.wikipedia.org/wiki/Notch_signaling_pathway
- ↑ https://www.cellsignal.com/contents/science-cst-pathways-stem-cell-markers/notch-signaling-interactive-pathway/pathways-notch
- ↑ https://www.ncbi.nlm.nih.gov/pubmed/?term=notch

