Difference between revisions of "Part:BBa K2627003"
| (One intermediate revision by one other user not shown) | |||
| Line 6: | Line 6: | ||
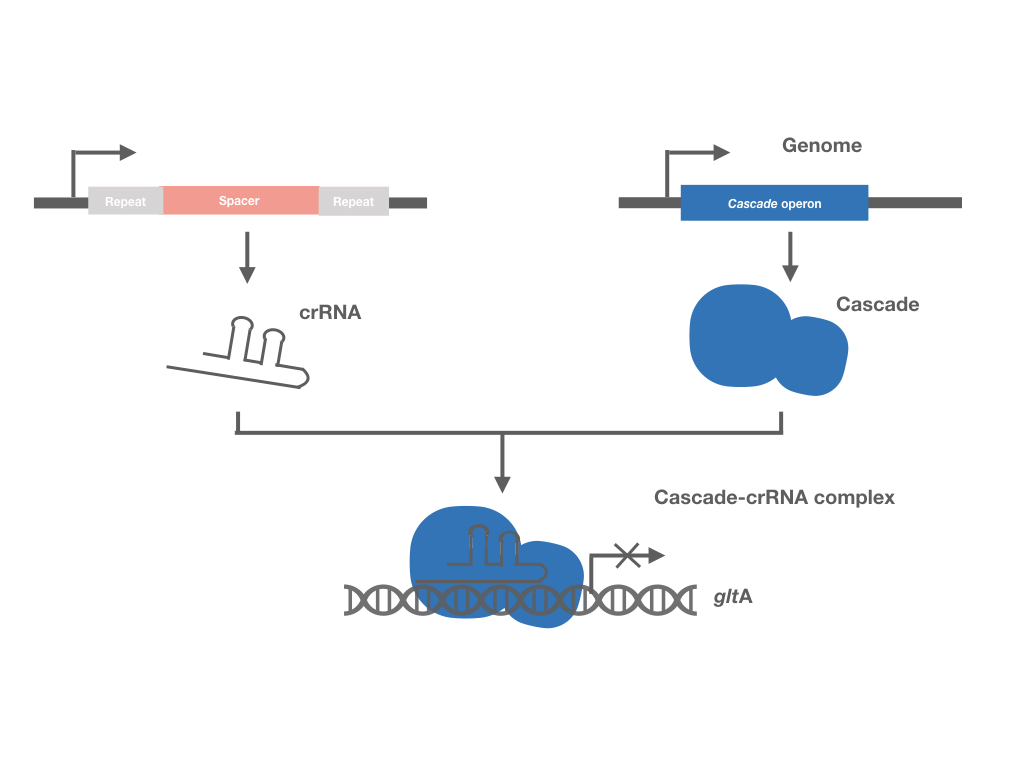
Similar to the other two CRISPR systems, crRNA in the Type I-E system sequences are recognized by ribonucleoprotein complex Cascade during target DNA binding. The ribonucleoprotein complex Cascade is composed of a 61 nt crRNA, and five different Cas proteins in an uneven stoichiometry: Cse11Cse22Cas76Cas51Cas6e1, encoded by 8 genes separately (cas3, cse1, cse2, cas7, cas5, cas6e, cas1, cas2). The interference function of this system requires Cas3, a large protein with nuclease and helicase activities. Therefore, by knocking out cas3 and activating the expression of CRISPR-associated complex for antiviral defense (Cascade), the CRISPR system is inactivated for its DNA-cutting function, with its DNA-binding function maintained, which can interference the expression of the target genes.</p> | Similar to the other two CRISPR systems, crRNA in the Type I-E system sequences are recognized by ribonucleoprotein complex Cascade during target DNA binding. The ribonucleoprotein complex Cascade is composed of a 61 nt crRNA, and five different Cas proteins in an uneven stoichiometry: Cse11Cse22Cas76Cas51Cas6e1, encoded by 8 genes separately (cas3, cse1, cse2, cas7, cas5, cas6e, cas1, cas2). The interference function of this system requires Cas3, a large protein with nuclease and helicase activities. Therefore, by knocking out cas3 and activating the expression of CRISPR-associated complex for antiviral defense (Cascade), the CRISPR system is inactivated for its DNA-cutting function, with its DNA-binding function maintained, which can interference the expression of the target genes.</p> | ||
| − | [[File:T--SDU-China-- | + | [[File:T--SDU-China--cascade.jpeg|500px|center]] |
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
| Line 19: | Line 19: | ||
<partinfo>BBa_K2627003 parameters</partinfo> | <partinfo>BBa_K2627003 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | |||
==Results== | ==Results== | ||
| + | ===Characterization of part BBa_K2627003=== | ||
<p>Part BBa_K2627003 was characterized by measuring bacteria growth curve when the cells were induced by light; arabinose respectively, and we found that the effect of this system was related to the copy number of plamid.</p> | <p>Part BBa_K2627003 was characterized by measuring bacteria growth curve when the cells were induced by light; arabinose respectively, and we found that the effect of this system was related to the copy number of plamid.</p> | ||
[[File:T--SDU-China--crrnar.png|800px|center|thumb|<p align="justify">'''Figure 1. CRISPRi system was induced by light. After the verification of the culture condition, we investigated the dynamic regulation effects of targeting gltA with light-controlled system through switching red light to green light at 2h, 5h and 9h. As shown in figure , all strains induced at different time grow better compared with illuminating at 0h. Bacteria growth was inhibited the most when being induced at 5h, showing an obvious palliation after sensing green light. The strains induced at the early-log phase (2h) grew slower than the control (upon red illumination), whereas it continuously grew and maintained a relatively high K value. It was supposed that cells growing at a moderate speed at early consumed less nutrition, thus drove the biomass constantly increase and had a higher level of K value. In accordance with the hypothesis, we concluded the negative control without crRNA targeting gltA which presented a lowest level of K value consumed too much nutrition so that it can’t satisfy the need of cell in late period of growth. In addition, inducing crRNA expression at 9h through green light, we found there were little growth variance between the strains green light induced at 9h and the control illuminated at red light continuously. It was supposed that at the stationary phase there was nearly no effect for inhibiting TCA cycle to repress the cell’s growth, which might due to its metabolic flow. The difference between the regulation effect of different induction time isn’t obvious through light switching compared with L-arabinose, might due to light sensor CcaS/CcaR system which only have 4.8-fold difference between red/green light induction.'''</p>]] | [[File:T--SDU-China--crrnar.png|800px|center|thumb|<p align="justify">'''Figure 1. CRISPRi system was induced by light. After the verification of the culture condition, we investigated the dynamic regulation effects of targeting gltA with light-controlled system through switching red light to green light at 2h, 5h and 9h. As shown in figure , all strains induced at different time grow better compared with illuminating at 0h. Bacteria growth was inhibited the most when being induced at 5h, showing an obvious palliation after sensing green light. The strains induced at the early-log phase (2h) grew slower than the control (upon red illumination), whereas it continuously grew and maintained a relatively high K value. It was supposed that cells growing at a moderate speed at early consumed less nutrition, thus drove the biomass constantly increase and had a higher level of K value. In accordance with the hypothesis, we concluded the negative control without crRNA targeting gltA which presented a lowest level of K value consumed too much nutrition so that it can’t satisfy the need of cell in late period of growth. In addition, inducing crRNA expression at 9h through green light, we found there were little growth variance between the strains green light induced at 9h and the control illuminated at red light continuously. It was supposed that at the stationary phase there was nearly no effect for inhibiting TCA cycle to repress the cell’s growth, which might due to its metabolic flow. The difference between the regulation effect of different induction time isn’t obvious through light switching compared with L-arabinose, might due to light sensor CcaS/CcaR system which only have 4.8-fold difference between red/green light induction.'''</p>]] | ||
[[File:T--SDU-China--crrnar2.png|800px|center|thumb|<p align="justify">'''Figure 2. CRISPRi system was induced by arabinose. With 0.2% L-arabinose induction, the samples showed an obvious repression on growth compared with the ones without L-arabinose (figure 2.). furthermore, we investigated the regulatory effects of targeting gltA at different stages of growth by adding L-arabinose at 0h, 2.5h, 5h, 7.5h and 9h. As shown in the figure, there presented an obvious difference between different induced time, in which the strains added L-arabinose at the beginning had the strongest repression in growth while with the delay of the induction time, the effect of inhibiting growth was gradually weakened. It indicated that this system can inhibit TCA cycle through inducing crRNA targeting gltA at different time, thus it proved that our idea that dynamically regulating metabolic flux was feasible to some extent.'''</p>]] | [[File:T--SDU-China--crrnar2.png|800px|center|thumb|<p align="justify">'''Figure 2. CRISPRi system was induced by arabinose. With 0.2% L-arabinose induction, the samples showed an obvious repression on growth compared with the ones without L-arabinose (figure 2.). furthermore, we investigated the regulatory effects of targeting gltA at different stages of growth by adding L-arabinose at 0h, 2.5h, 5h, 7.5h and 9h. As shown in the figure, there presented an obvious difference between different induced time, in which the strains added L-arabinose at the beginning had the strongest repression in growth while with the delay of the induction time, the effect of inhibiting growth was gradually weakened. It indicated that this system can inhibit TCA cycle through inducing crRNA targeting gltA at different time, thus it proved that our idea that dynamically regulating metabolic flux was feasible to some extent.'''</p>]] | ||
[[File:T--SDU-China--crrnar3.png|800px|center|thumb|<p align="justify">'''Figure 3. Effects of copy number. To optimize the expression of crRNAs in EE-E15, then we constructed a medium-copy plasmid (pSR43.6) and high-copy plasmid (phz) to express crRNA targeting endogenous gltA. The two strains showed significant variance in growth in figure 4, suggesting crRNA on high-copy plasmid showed stronger repression than on medium-copy one. We supposed that the inhibition function was related to the quantity of crRNA and light-sensing promotor PcpG2-172 works well in high-copy number, leading to this result. It indicated that copy-number for light-induced crRNA had a great impact on this photoactivated regulation system.'''</p>]] | [[File:T--SDU-China--crrnar3.png|800px|center|thumb|<p align="justify">'''Figure 3. Effects of copy number. To optimize the expression of crRNAs in EE-E15, then we constructed a medium-copy plasmid (pSR43.6) and high-copy plasmid (phz) to express crRNA targeting endogenous gltA. The two strains showed significant variance in growth in figure 4, suggesting crRNA on high-copy plasmid showed stronger repression than on medium-copy one. We supposed that the inhibition function was related to the quantity of crRNA and light-sensing promotor PcpG2-172 works well in high-copy number, leading to this result. It indicated that copy-number for light-induced crRNA had a great impact on this photoactivated regulation system.'''</p>]] | ||
Latest revision as of 03:56, 18 October 2018
crRNA targeting GltA
Usage and Biology
Clustered regularly interspaced short palindromic repeats (CRISPR) is a family of DNA sequences found within the genomes of prokaryotic organisms such as bacteria and archaea, which have the function in binding and cutting DNA. There are three types of CRISPR system found in microorganisms to date: type I, type II and type III. In Type I and Type III systems the long precursor CRISPR RNA (pre-crRNA) is processed by CRISPR specific endoribonucleases into small CRISPR RNAs (crRNAs) that contain a repeat sequence flaked by portions of the adjacent CRISPR repeat sequence. In contrast, the pre-crRNA in Type II systems is processed by RNase III.
Similar to the other two CRISPR systems, crRNA in the Type I-E system sequences are recognized by ribonucleoprotein complex Cascade during target DNA binding. The ribonucleoprotein complex Cascade is composed of a 61 nt crRNA, and five different Cas proteins in an uneven stoichiometry: Cse11Cse22Cas76Cas51Cas6e1, encoded by 8 genes separately (cas3, cse1, cse2, cas7, cas5, cas6e, cas1, cas2). The interference function of this system requires Cas3, a large protein with nuclease and helicase activities. Therefore, by knocking out cas3 and activating the expression of CRISPR-associated complex for antiviral defense (Cascade), the CRISPR system is inactivated for its DNA-cutting function, with its DNA-binding function maintained, which can interference the expression of the target genes.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Results
Characterization of part BBa_K2627003
Part BBa_K2627003 was characterized by measuring bacteria growth curve when the cells were induced by light; arabinose respectively, and we found that the effect of this system was related to the copy number of plamid.
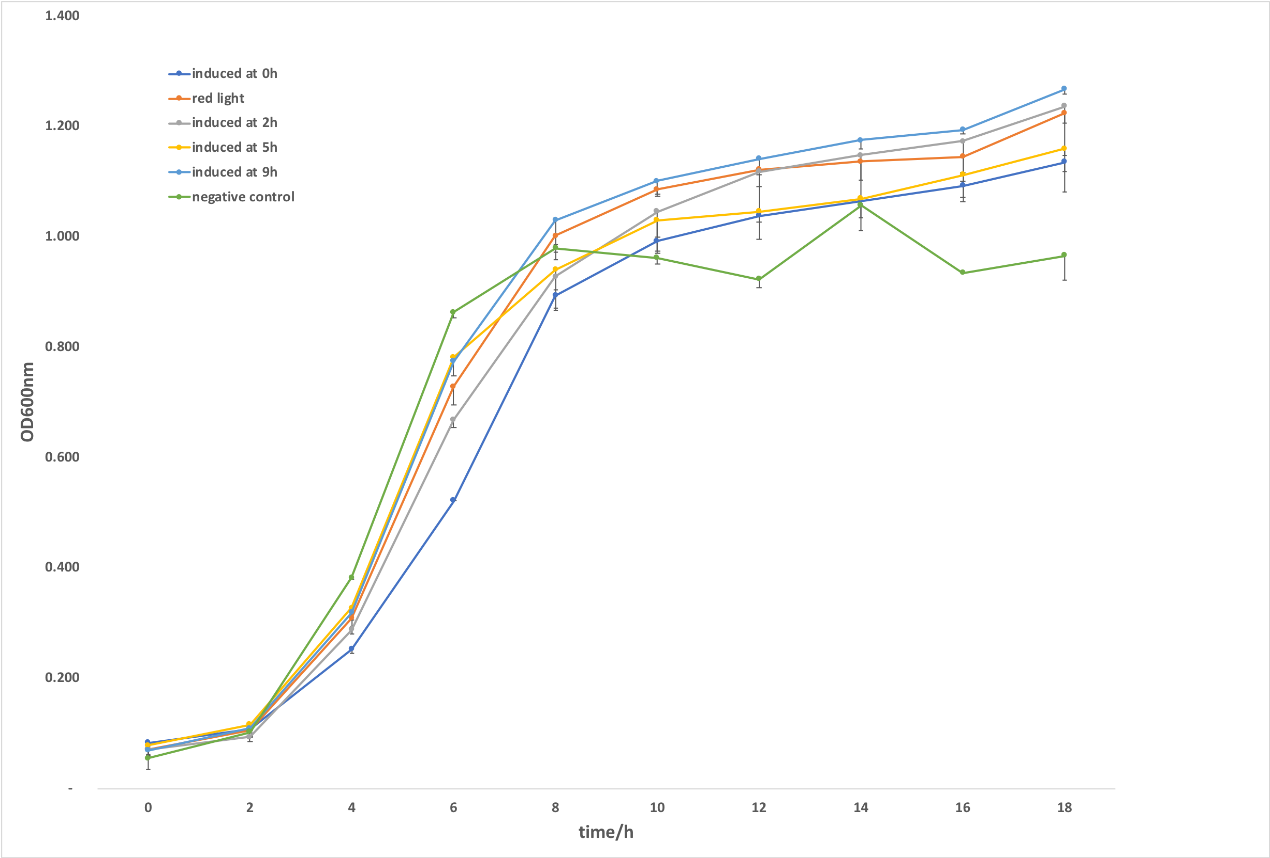
Figure 1. CRISPRi system was induced by light. After the verification of the culture condition, we investigated the dynamic regulation effects of targeting gltA with light-controlled system through switching red light to green light at 2h, 5h and 9h. As shown in figure , all strains induced at different time grow better compared with illuminating at 0h. Bacteria growth was inhibited the most when being induced at 5h, showing an obvious palliation after sensing green light. The strains induced at the early-log phase (2h) grew slower than the control (upon red illumination), whereas it continuously grew and maintained a relatively high K value. It was supposed that cells growing at a moderate speed at early consumed less nutrition, thus drove the biomass constantly increase and had a higher level of K value. In accordance with the hypothesis, we concluded the negative control without crRNA targeting gltA which presented a lowest level of K value consumed too much nutrition so that it can’t satisfy the need of cell in late period of growth. In addition, inducing crRNA expression at 9h through green light, we found there were little growth variance between the strains green light induced at 9h and the control illuminated at red light continuously. It was supposed that at the stationary phase there was nearly no effect for inhibiting TCA cycle to repress the cell’s growth, which might due to its metabolic flow. The difference between the regulation effect of different induction time isn’t obvious through light switching compared with L-arabinose, might due to light sensor CcaS/CcaR system which only have 4.8-fold difference between red/green light induction.
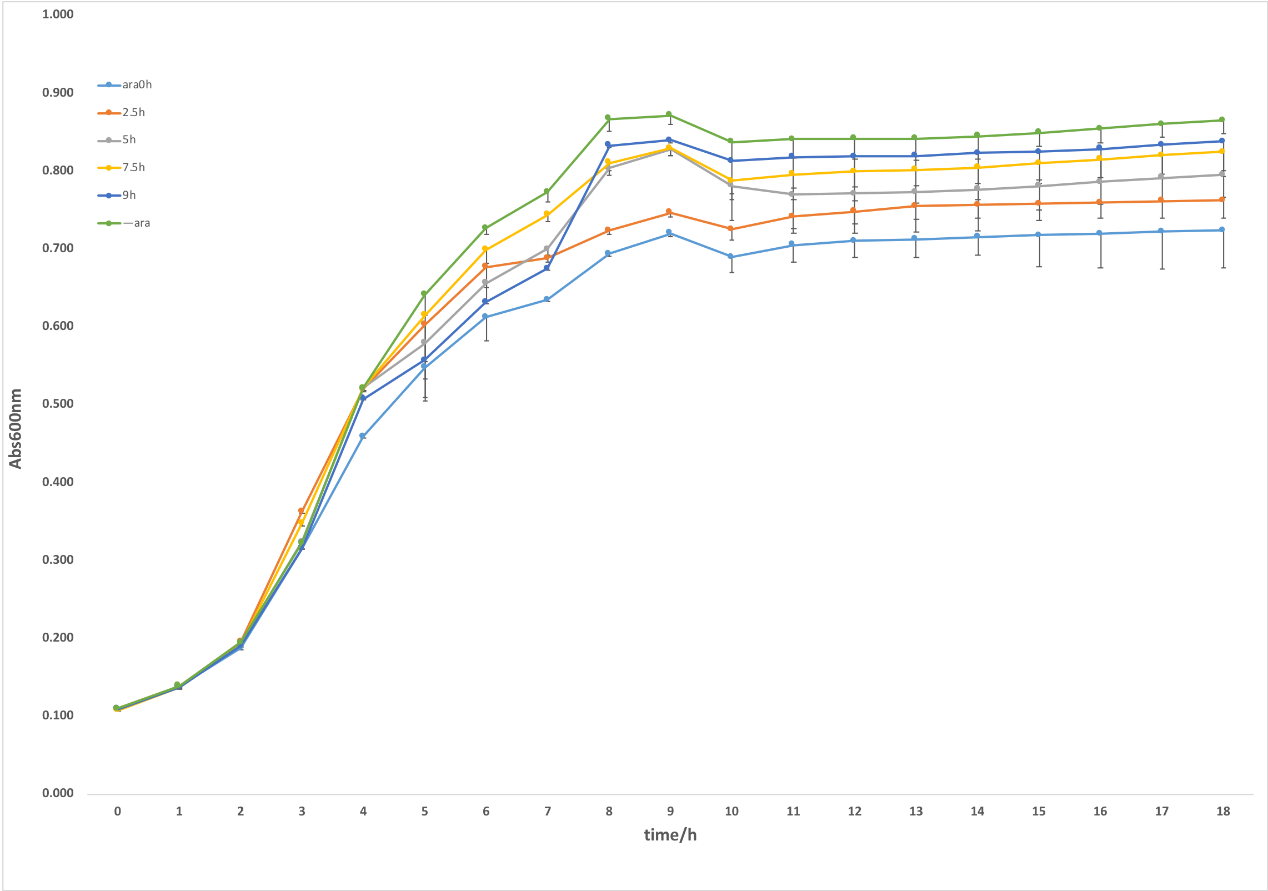
Figure 2. CRISPRi system was induced by arabinose. With 0.2% L-arabinose induction, the samples showed an obvious repression on growth compared with the ones without L-arabinose (figure 2.). furthermore, we investigated the regulatory effects of targeting gltA at different stages of growth by adding L-arabinose at 0h, 2.5h, 5h, 7.5h and 9h. As shown in the figure, there presented an obvious difference between different induced time, in which the strains added L-arabinose at the beginning had the strongest repression in growth while with the delay of the induction time, the effect of inhibiting growth was gradually weakened. It indicated that this system can inhibit TCA cycle through inducing crRNA targeting gltA at different time, thus it proved that our idea that dynamically regulating metabolic flux was feasible to some extent.
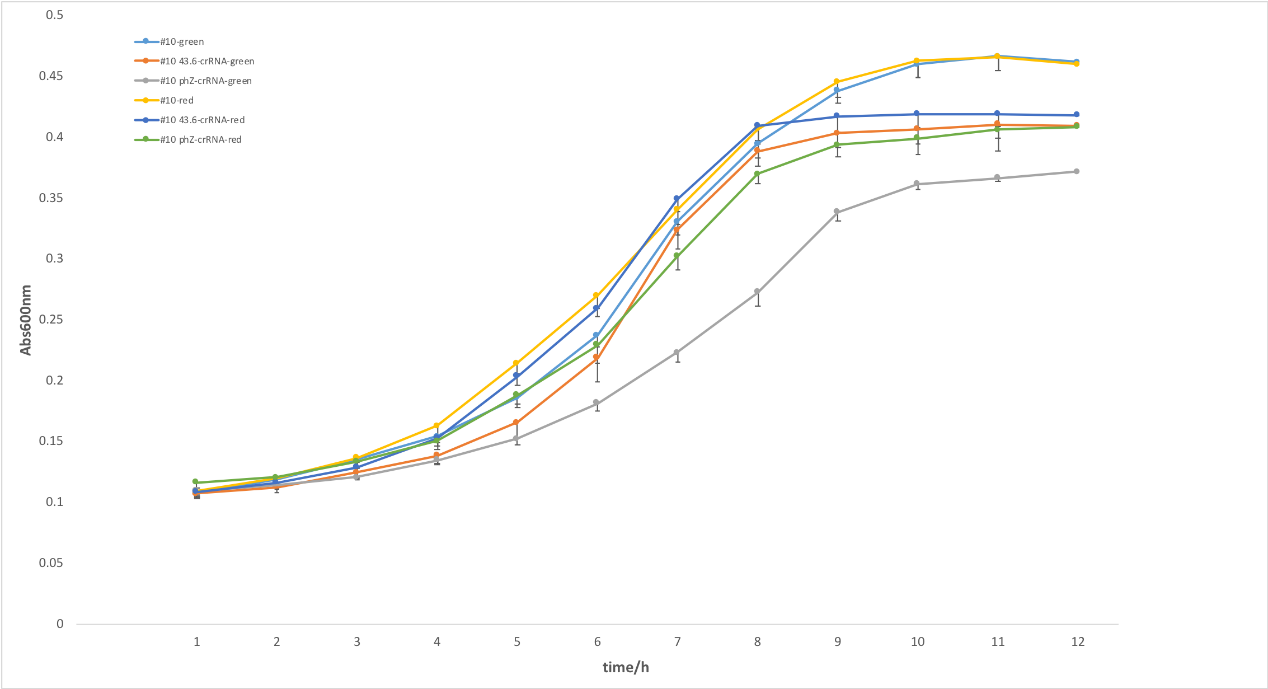
Figure 3. Effects of copy number. To optimize the expression of crRNAs in EE-E15, then we constructed a medium-copy plasmid (pSR43.6) and high-copy plasmid (phz) to express crRNA targeting endogenous gltA. The two strains showed significant variance in growth in figure 4, suggesting crRNA on high-copy plasmid showed stronger repression than on medium-copy one. We supposed that the inhibition function was related to the quantity of crRNA and light-sensing promotor PcpG2-172 works well in high-copy number, leading to this result. It indicated that copy-number for light-induced crRNA had a great impact on this photoactivated regulation system.