Difference between revisions of "Part:BBa K1789001"
Emilywatson (Talk | contribs) |
Emilywatson (Talk | contribs) |
||
| (5 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
<partinfo>BBa_K1789001 short</partinfo> | <partinfo>BBa_K1789001 short</partinfo> | ||
| − | This part is the coding sequence of the enzyme Indoleacetimide hydrolase | + | This part is the coding sequence of the enzyme Indoleacetimide hydrolase IaaH. This enzyme catalyses the hydrolysis of indoleacetamide to indoleacetate and ammonia. This part is gathered from the 2-step pathway in Pseudomonas savastanoi producing the auxin, indole-3-acetic acid. |
==Usage and Biology== | ==Usage and Biology== | ||
| Line 73: | Line 73: | ||
'''iGEM18_UNSW_Australia:''' | '''iGEM18_UNSW_Australia:''' | ||
<p>'''His IaaH-SpyTag''' <partinfo>BBa_K2710005</partinfo> </p> | <p>'''His IaaH-SpyTag''' <partinfo>BBa_K2710005</partinfo> </p> | ||
| − | The 2018 UNSW iGEM team designed a new IaaH encoding part for improved functionality of the IaaH enzyme. A HisTag and GSG linker were added to the N-terminus of the enzyme, and a SpyTag and GSG linker was added to the C-terminus. Following these additions, the team was able to successfully express and purify the new part utilising the HisTag's affinity for nickel ions to purify the enzyme using IMAC purification. We also demonstrated the ability of IaaH with SpyTag to covalently bind with SpyCatcher proteins through SDS-PAGE. | + | The 2018 UNSW iGEM team designed a new IaaH encoding part for improved functionality of the IaaH enzyme. A HisTag and GSG linker were added to the N-terminus of the enzyme, and a SpyTag and GSG linker was added to the C-terminus. Following these additions, the team was able to successfully express and purify the new part utilising the HisTag's affinity for nickel ions to purify the enzyme using IMAC purification <b>(Figure 4)</b>. We also demonstrated the ability of IaaH with SpyTag to covalently bind with SpyCatcher proteins through SDS-PAGE <b>(Figure 5)</b>. |
| + | |||
| + | <br /> | ||
| + | https://static.igem.org/mediawiki/parts/thumb/5/58/T--UNSW_Australia--IaaH-purifications.jpeg/800px-T--UNSW_Australia--IaaH-purifications.jpeg | ||
| + | |||
| + | <b>Figure 4:</b> SDS-PAGE analysis of IMAC purification of IaaH (BBa_K1789001) and His-IaaH-SpyTag (BBa_K2710005). SeeBlue Plus 2 Pre-stained Protein Standard (Invitrogen) was used as the molecular weight standard. Lanes are labelled as flow through (FT), wash (W) and elutions (E1 and E2). The IaaH enzyme encoded by BBa_K1789001 was not His-tagged and thus unable to be purified by IMAC as seen by the absence of a distinct band at the expected molecular weight range (49 kDa). In contrast, the improved His-IaaH-SpyTag was successfully purified as reflected by the clear bands seen at the expected molecular weight range (53 kDa) in the elution lanes. | ||
| + | <br /> | ||
| + | |||
| + | https://static.igem.org/mediawiki/2018/thumb/b/b7/T--UNSW_Australia--assembly3.jpeg/291px-T--UNSW_Australia--assembly3.jpeg | ||
| + | <p class="figure-legend fig4-leg"><b>Figure 5:</b> aPFD-SpyC covalently attaches to IaaH-SpyT. The bands indicating successful attachment of IaaH-SpyT to aPFD-SpyC are boxed in red. </p> | ||
Latest revision as of 03:17, 18 October 2018
IaaH
This part is the coding sequence of the enzyme Indoleacetimide hydrolase IaaH. This enzyme catalyses the hydrolysis of indoleacetamide to indoleacetate and ammonia. This part is gathered from the 2-step pathway in Pseudomonas savastanoi producing the auxin, indole-3-acetic acid.
Usage and Biology
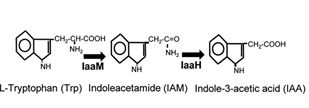
Indole-3-acetic acid (IAA), also known as auxin, is a plant hormone, which can promote growth and protect the soil from erosion. IAA can be synthesized through IAM pathway, originated from Pseudomonas savastanoi. Two important enzymes are contained in this pathway, AKA IaaM and IaaH.
IaaM is the tryptophan-2-mono-oxygenase. This enzyme catalyzes the oxidative carboxylation of L-tryptophan to indole-3-acetamide. IaaH is the indoleacetimide hydrolase. This enzyme catalyses the hydrolysis of indoleacetamide to indoleacetate and ammonia. This pathway is relevantly easy to be expressed in prokaryotic cells, and its substrate (Trp) can be synthesized by host cells.
Sequence and Features
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 1005
- 1000COMPATIBLE WITH RFC[1000]
Experimental Validation
This part is validated through four ways: enzyme cutting, PCR, Sequence, and functional testing
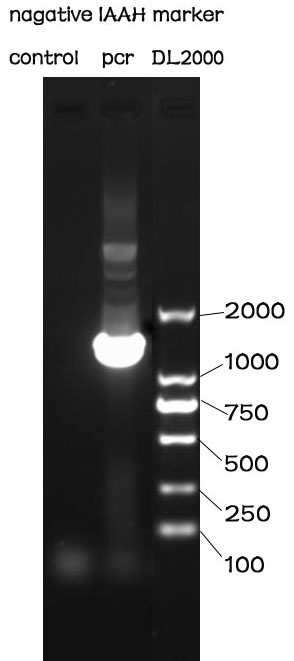
PCR
Methods
The PCR is performed with Premix EX Taq by Takara.
F-Prime: 5’- GGAATTCGCGGCCGCTTCTAGAGATGCGCGAAATG -3’
R-Prime: 5’- GCGGGCGGCGGACTAGTCTTATTAGCCTTTTAACAC -3’
The PCR protocol is selected based on the Users Manuel. The Electrophoresis was performed on a 1% Agarose glu.
Results
The result of the agarose electrophoresis was shown on the picture above.
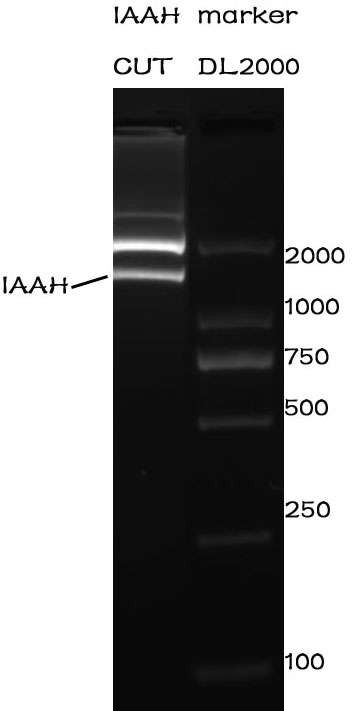
Enzyme cutting
Methods
After the assembly ,the plasmid was transferred into the Competent E. coli DH5α). After culturing overnight in LB,we minipreped the plasmid for cutting. The preparation of the plasmid was performed with TIANprep Mini Plasmid Kit from TIANGEN. The cutting procedure was performed with EcoRI and XbaI restriction endonuclease bought from TAKARA.
The plasmid was cutted in a 20μL system at 37 ℃ for 2 hours. The Electrophoresis was performed on a 1% Agarose glu.
Results
The result of the agarose electrophoresis was shown on the picture above.
Functional Test
This part is tested together with the part BBa_K1789000, in the device BBa_K1789022.
The result shows that this part can work as expected to generate IAA together with IAAM.
Improved Design
iGEM18_UNSW_Australia:
His IaaH-SpyTag BBa_K2710005
The 2018 UNSW iGEM team designed a new IaaH encoding part for improved functionality of the IaaH enzyme. A HisTag and GSG linker were added to the N-terminus of the enzyme, and a SpyTag and GSG linker was added to the C-terminus. Following these additions, the team was able to successfully express and purify the new part utilising the HisTag's affinity for nickel ions to purify the enzyme using IMAC purification (Figure 4). We also demonstrated the ability of IaaH with SpyTag to covalently bind with SpyCatcher proteins through SDS-PAGE (Figure 5).

Figure 4: SDS-PAGE analysis of IMAC purification of IaaH (BBa_K1789001) and His-IaaH-SpyTag (BBa_K2710005). SeeBlue Plus 2 Pre-stained Protein Standard (Invitrogen) was used as the molecular weight standard. Lanes are labelled as flow through (FT), wash (W) and elutions (E1 and E2). The IaaH enzyme encoded by BBa_K1789001 was not His-tagged and thus unable to be purified by IMAC as seen by the absence of a distinct band at the expected molecular weight range (49 kDa). In contrast, the improved His-IaaH-SpyTag was successfully purified as reflected by the clear bands seen at the expected molecular weight range (53 kDa) in the elution lanes.

Figure 5: aPFD-SpyC covalently attaches to IaaH-SpyT. The bands indicating successful attachment of IaaH-SpyT to aPFD-SpyC are boxed in red.