Difference between revisions of "Part:BBa K2549041"
(→Biology) |
(→Our characterization) |
||
| (2 intermediate revisions by the same user not shown) | |||
| Line 4: | Line 4: | ||
This part is one of the downstream elements of our amplifier. It is constructed by fusing NLS ([[Part:BBa_K2549054]]) and TEV protease ([[Part:BBa_K2549013]]), from N terminal to C terminal. NLS is a short nuclear location sequence from SV40 large T antigen. TEVp is a mutant TEV (tobacco etch virus) protease whose autoactivation is removed, thus performing more efficient cleavage<ref>Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Kapust RB, Tözsér J, Fox JD, ..., Copeland TD, Waugh DS. Protein Eng, 2001 Dec;14(12):993-1000 PMID: 11809930</ref>. When coexpressed with VP64-dNLS-ZF21.16 ([[Part:BBa_K2549039]]) in the same cell, it can destroy the nuclear localization sequence and prevent the transcription activator from inducing the gene expression. Similarly, the cleavage can be conducted with the KRAB-dNLS-ZF21.16 ([[Part:BBa_K2549040]]), thus keeping the repressor from switching off the gene expression. | This part is one of the downstream elements of our amplifier. It is constructed by fusing NLS ([[Part:BBa_K2549054]]) and TEV protease ([[Part:BBa_K2549013]]), from N terminal to C terminal. NLS is a short nuclear location sequence from SV40 large T antigen. TEVp is a mutant TEV (tobacco etch virus) protease whose autoactivation is removed, thus performing more efficient cleavage<ref>Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Kapust RB, Tözsér J, Fox JD, ..., Copeland TD, Waugh DS. Protein Eng, 2001 Dec;14(12):993-1000 PMID: 11809930</ref>. When coexpressed with VP64-dNLS-ZF21.16 ([[Part:BBa_K2549039]]) in the same cell, it can destroy the nuclear localization sequence and prevent the transcription activator from inducing the gene expression. Similarly, the cleavage can be conducted with the KRAB-dNLS-ZF21.16 ([[Part:BBa_K2549040]]), thus keeping the repressor from switching off the gene expression. | ||
| + | |||
| + | <!-- --> | ||
| + | <span class='h3bb'>Sequence and Features</span> | ||
| + | <partinfo>BBa_K2549041 SequenceAndFeatures</partinfo> | ||
| + | |||
<!-- Add more about the biology of this part here --> | <!-- Add more about the biology of this part here --> | ||
===Biology=== | ===Biology=== | ||
| + | =====Our characterization===== | ||
| + | [[File:IMPLY.png|none|410px|thumb|'''TEVp-based IMPLY gate.''' A degradable EGFP (d2EGFP) is produced downstream the promoter of the Combiner to indicate the output (CusP) strength. DBD, DNA binding domain which is zinc finger in our assay. SD, silencing-form transcriptional domain. RE, responsive elements. RFI, relative fluorescence intensity. More details please visit http://2018.igem.org/Team:Fudan/Results and http://2018.igem.org/Team:Fudan/Measurement .]] | ||
| + | |||
| + | We show that when KRAB-dNLS-ZF21.16 ([[Part:BBa_K2549040]]) exist while TEVp ([[Part:BBa_K2549041]]) did not, the expression of d2EGFP was repressed by KRAB. When TEVp and KRAB-dNLS-ZF21.16 both were present, TEVp cleaves dNLS and prevents KRAB functional, although the release from the suppression was not as strong as we hoped. NS3 cis-protease is on our list to try next<ref>Chemogenetic control of gene expression and cell signaling with antiviral drugs. Tague EP, Dotson HL, Tunney SN, Sloas DC, Ngo JT. Nat Methods, 2018 Jul;15(7):519-522 PMID: 29967495; DOI: 10.1038/s41592-018-0042-y</ref>. | ||
| + | |||
====TEV protease-based transcription regulation or proteolysis-only activity==== | ====TEV protease-based transcription regulation or proteolysis-only activity==== | ||
TEV protease is widely used in synthetic biology for its high cleavage specificity (targetting amino acids sequence ENLYFQG/S between QG or QS)<ref>Release of proteins and peptides from fusion proteins using a recombinant plant virus proteinase. Parks TD, Leuther KK, Howard ED, Johnston SA, Dougherty WG. Anal Biochem, 1994 Feb;216(2):413-7 PMID: 8179197; DOI: 10.1006/abio.1994.1060</ref>. Rossner MJ et al have demonstrated the TEV activity-dependent activation of several reporters<ref>Monitoring regulated protein-protein interactions using split TEV. Wehr MC, Laage R, Bolz U, ..., Nave KA, Rossner MJ. Nat Methods, 2006 Dec;3(12):985-93 PMID: 17072307; DOI: 10.1038/nmeth967</ref>. '''In our system, we set the TEV protease specific cleavage site into the spacer region of the nuclear localization sequence<ref>Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Robbins J, Dilworth SM, Laskey RA, Dingwall C. Cell, 1991 Feb;64(3):615-23 PMID: 1991323</ref>, which is the core design of the TEV protease-based complex logic gates.''' | TEV protease is widely used in synthetic biology for its high cleavage specificity (targetting amino acids sequence ENLYFQG/S between QG or QS)<ref>Release of proteins and peptides from fusion proteins using a recombinant plant virus proteinase. Parks TD, Leuther KK, Howard ED, Johnston SA, Dougherty WG. Anal Biochem, 1994 Feb;216(2):413-7 PMID: 8179197; DOI: 10.1006/abio.1994.1060</ref>. Rossner MJ et al have demonstrated the TEV activity-dependent activation of several reporters<ref>Monitoring regulated protein-protein interactions using split TEV. Wehr MC, Laage R, Bolz U, ..., Nave KA, Rossner MJ. Nat Methods, 2006 Dec;3(12):985-93 PMID: 17072307; DOI: 10.1038/nmeth967</ref>. '''In our system, we set the TEV protease specific cleavage site into the spacer region of the nuclear localization sequence<ref>Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Robbins J, Dilworth SM, Laskey RA, Dingwall C. Cell, 1991 Feb;64(3):615-23 PMID: 1991323</ref>, which is the core design of the TEV protease-based complex logic gates.''' | ||
| Line 19: | Line 29: | ||
Flow cytometry results indicate that when KRAB-dNLS-ZF21.16 exists while TEVp doesn't exist that the expression level of d2EGFP is repressed. When TEVp and KRAB-dNLS-ZF21.16 coexist, TEVp cleaves dNLS and prevents KRAB from entering into the nucleus. Thus the repression is not severe as when KRAB-dNLS-ZF21.16 is present while TEVp is absent. | Flow cytometry results indicate that when KRAB-dNLS-ZF21.16 exists while TEVp doesn't exist that the expression level of d2EGFP is repressed. When TEVp and KRAB-dNLS-ZF21.16 coexist, TEVp cleaves dNLS and prevents KRAB from entering into the nucleus. Thus the repression is not severe as when KRAB-dNLS-ZF21.16 is present while TEVp is absent. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
Latest revision as of 21:34, 17 October 2018
NLS-TEVp
This part is one of the downstream elements of our amplifier. It is constructed by fusing NLS (Part:BBa_K2549054) and TEV protease (Part:BBa_K2549013), from N terminal to C terminal. NLS is a short nuclear location sequence from SV40 large T antigen. TEVp is a mutant TEV (tobacco etch virus) protease whose autoactivation is removed, thus performing more efficient cleavage[1]. When coexpressed with VP64-dNLS-ZF21.16 (Part:BBa_K2549039) in the same cell, it can destroy the nuclear localization sequence and prevent the transcription activator from inducing the gene expression. Similarly, the cleavage can be conducted with the KRAB-dNLS-ZF21.16 (Part:BBa_K2549040), thus keeping the repressor from switching off the gene expression.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 337
Biology
Our characterization
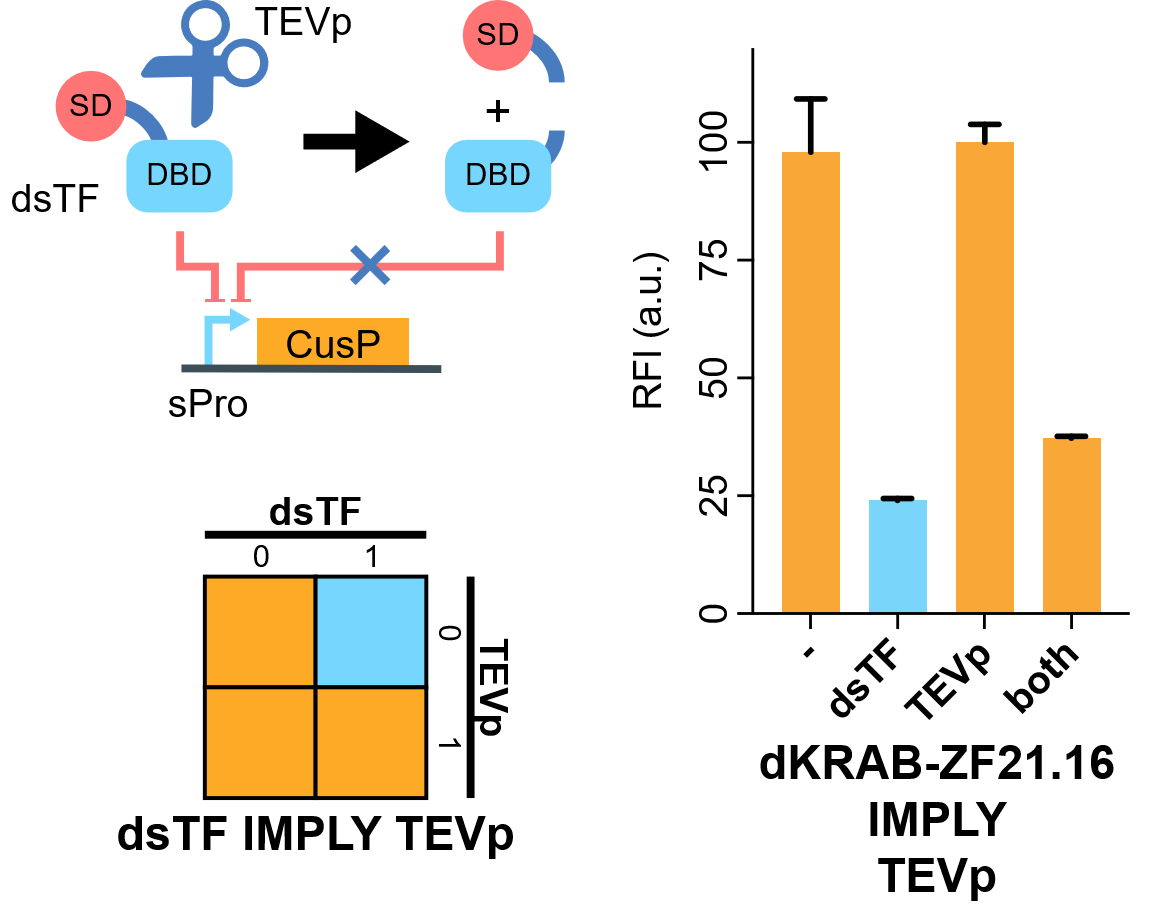
We show that when KRAB-dNLS-ZF21.16 (Part:BBa_K2549040) exist while TEVp (Part:BBa_K2549041) did not, the expression of d2EGFP was repressed by KRAB. When TEVp and KRAB-dNLS-ZF21.16 both were present, TEVp cleaves dNLS and prevents KRAB functional, although the release from the suppression was not as strong as we hoped. NS3 cis-protease is on our list to try next[2].
TEV protease-based transcription regulation or proteolysis-only activity
TEV protease is widely used in synthetic biology for its high cleavage specificity (targetting amino acids sequence ENLYFQG/S between QG or QS)[3]. Rossner MJ et al have demonstrated the TEV activity-dependent activation of several reporters[4]. In our system, we set the TEV protease specific cleavage site into the spacer region of the nuclear localization sequence[5], which is the core design of the TEV protease-based complex logic gates.
For more details about TEV protease, please refer to our TEVp (Part:BBa_K2549013).
Characterization
It works as we designed.

Flow cytometry results indicate that when KRAB-dNLS-ZF21.16 exists while TEVp doesn't exist that the expression level of d2EGFP is repressed. When TEVp and KRAB-dNLS-ZF21.16 coexist, TEVp cleaves dNLS and prevents KRAB from entering into the nucleus. Thus the repression is not severe as when KRAB-dNLS-ZF21.16 is present while TEVp is absent.
References
- ↑ Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Kapust RB, Tözsér J, Fox JD, ..., Copeland TD, Waugh DS. Protein Eng, 2001 Dec;14(12):993-1000 PMID: 11809930
- ↑ Chemogenetic control of gene expression and cell signaling with antiviral drugs. Tague EP, Dotson HL, Tunney SN, Sloas DC, Ngo JT. Nat Methods, 2018 Jul;15(7):519-522 PMID: 29967495; DOI: 10.1038/s41592-018-0042-y
- ↑ Release of proteins and peptides from fusion proteins using a recombinant plant virus proteinase. Parks TD, Leuther KK, Howard ED, Johnston SA, Dougherty WG. Anal Biochem, 1994 Feb;216(2):413-7 PMID: 8179197; DOI: 10.1006/abio.1994.1060
- ↑ Monitoring regulated protein-protein interactions using split TEV. Wehr MC, Laage R, Bolz U, ..., Nave KA, Rossner MJ. Nat Methods, 2006 Dec;3(12):985-93 PMID: 17072307; DOI: 10.1038/nmeth967
- ↑ Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Robbins J, Dilworth SM, Laskey RA, Dingwall C. Cell, 1991 Feb;64(3):615-23 PMID: 1991323
