Difference between revisions of "Part:BBa K2856003"
(→Usage and Biology) |
|||
| (7 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K2856003 short</partinfo> | <partinfo>BBa_K2856003 short</partinfo> | ||
| + | <br><br> | ||
| − | BBa_K2856003 harbors an adhesion-associated protein called cwaA. The cwaA encodes a protein containing multiple domains, including five cell wall surface anchor repeat domains and an LPxTG-like cell wall anchor motif. | + | BBa_K2856003 harbors an adhesion-associated protein called cwaA. The cwaA encodes a protein containing multiple domains, including five cell wall surface anchor repeat domains and an LPxTG-like cell wall anchor motif.<br> |
===Usage and Biology=== | ===Usage and Biology=== | ||
| + | <br> | ||
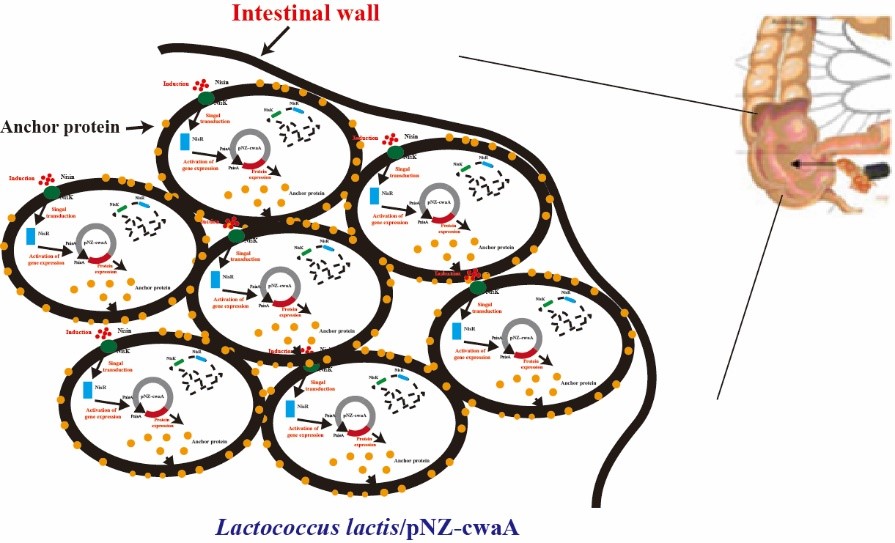
| + | CwaA protein encoded by gene cwaA is a cell wall-anchored protein. The C terminus of CwaA contains an LPQTDE (LPxTG-like cell wall anchoring) motif belonging to the gram-positive LPxTG anchor superfamily. Aside from the hexapeptide motif at the C terminus, CwaA possesses five cell wall surface anchor repeat domains. The specific hit domains of CwaA included epiglycanin (tandem-repeating region of mucin), OmpC (outer membrane protein), PT (the tetrapeptide XPTX repeat). Taken together, CwaA is a multidomain-containing cell wall-anchored protein that is very likely involved in cell adhesion. Thus, by expressing CwaA protein might be useful for improving the adhesivity of strains.<br>[http://2018.igem.org/Team:H14Z1_Hangzhou Team H14Z1_Hangzhou 2018]<br><br> | ||
| − | + | [[File:T--H14Z1 Hangzhou--Function_cwaA.jpeg|400px|thumb|centre| <p>'''Figure. 1 The function of protein CwaA in L. lactis.'''</p>]]<br><br> | |
| − | + | ||
| − | [[File:T--H14Z1 Hangzhou--Function_cwaA.jpeg| | + | |
===Construction and validation of plasmid pNZ-cwaA=== | ===Construction and validation of plasmid pNZ-cwaA=== | ||
| − | + | <br> | |
| − | Gene cwaA is from genomic DNA of Lactobacillus plantarum NL42 .In order to remove some illegal restriction enzyme sites and add a HIS tag, the gene cwaA was synthesized by Shanghai Generay Biotech (Shanghai, China). The gene cut with restriction enzyme Hind III and NcoI was ligased with plasmid pNZ8148 cut with the same enzyme. Then the ligation product was transferred to E.coli and spread on plates containing 10 mg/L chloramphenicol. Colonies on the plates were randomly picked and inoculated in 1ml LB medium for 3 hours at 37℃, 200 rpm. 1 μl culture were added to the PCR system as template. As shown in Figure. 2, all the picked colonies had the brand of cwaA, illustrating that the plasmid pNZ-cwaA was successfully constructed. | + | Gene cwaA is from genomic DNA of Lactobacillus plantarum NL42 .In order to remove some illegal restriction enzyme sites and add a HIS tag, the gene cwaA was synthesized by Shanghai Generay Biotech (Shanghai, China). The gene cut with restriction enzyme Hind III and NcoI was ligased with plasmid pNZ8148 cut with the same enzyme. Then the ligation product was transferred to E.coli and spread on plates containing 10 mg/L chloramphenicol. Colonies on the plates were randomly picked and inoculated in 1ml LB medium for 3 hours at 37℃, 200 rpm. 1 μl culture were added to the PCR system as template. As shown in Figure. 2, all the picked colonies had the brand of cwaA, illustrating that the plasmid pNZ-cwaA was successfully constructed.<br><br> |
| − | [[File:T--H14Z1 Hangzhou--plasmid_cwaA.jpeg|600px|thumb|centre| <p>'''Figure. 2 Validation of plasmid pNZ-cwaA. M represented marker. 1-4 represented three randomly picked colonies.'''</p>]] | + | [[File:T--H14Z1 Hangzhou--plasmid_cwaA.jpeg|600px|thumb|centre| <p>'''Figure. 2 Validation of plasmid pNZ-cwaA. M represented marker. 1-4 represented three randomly picked colonies.'''</p>]]<br><br> |
===Validation of autoaggregation of the recombinant L. lactis/pNZ-cwaA=== | ===Validation of autoaggregation of the recombinant L. lactis/pNZ-cwaA=== | ||
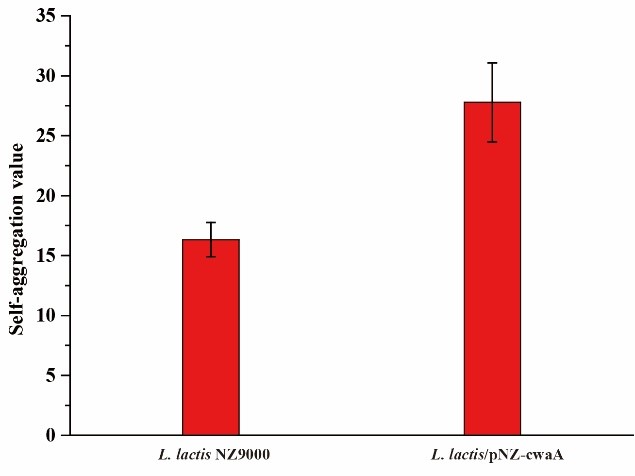
| − | To evaluate whether CwaA was involved in adhesion, we performed autoaggregation assays. The cells were collected and washed twice with o.1M PBS (pH=7.0).Then the cells were resuspended in 4ml PBS buffer with an initial OD600=0.5(A0) and the final OD600(At) of the upper most layer was measured after 4 hour cultivation in 30℃ without shaking. | + | <br> |
| − | + | To evaluate whether CwaA was involved in adhesion, we performed autoaggregation assays. The cells were collected and washed twice with o.1M PBS (pH=7.0).Then the cells were resuspended in 4ml PBS buffer with an initial OD600=0.5(A0) and the final OD600(At) of the upper most layer was measured after 4 hour cultivation in 30℃ without shaking.<br> | |
| − | Autoaggregation (%)=100-(At/A0)X100 | + | <ul> |
| − | [[File:T--H14Z1 Hangzhou--autoaggregation_cwaA.jpeg| | + | Autoaggregation (%)=100-(At/A0)X100<br><br> |
| + | [[File:T--H14Z1 Hangzhou--autoaggregation_cwaA.jpeg|400px|thumb|centre| <p>'''Figure. 3 Autoaggregation value of L. lactis NZ9000 and L. lactis/pNZ-cwaA.'''</p>]]<br><br> | ||
<!-- --> | <!-- --> | ||
Latest revision as of 12:19, 17 October 2018
Modification of cell wall anchor domain-containing protein (CwaA)
BBa_K2856003 harbors an adhesion-associated protein called cwaA. The cwaA encodes a protein containing multiple domains, including five cell wall surface anchor repeat domains and an LPxTG-like cell wall anchor motif.
Usage and Biology
CwaA protein encoded by gene cwaA is a cell wall-anchored protein. The C terminus of CwaA contains an LPQTDE (LPxTG-like cell wall anchoring) motif belonging to the gram-positive LPxTG anchor superfamily. Aside from the hexapeptide motif at the C terminus, CwaA possesses five cell wall surface anchor repeat domains. The specific hit domains of CwaA included epiglycanin (tandem-repeating region of mucin), OmpC (outer membrane protein), PT (the tetrapeptide XPTX repeat). Taken together, CwaA is a multidomain-containing cell wall-anchored protein that is very likely involved in cell adhesion. Thus, by expressing CwaA protein might be useful for improving the adhesivity of strains.
[http://2018.igem.org/Team:H14Z1_Hangzhou Team H14Z1_Hangzhou 2018]
Construction and validation of plasmid pNZ-cwaA
Gene cwaA is from genomic DNA of Lactobacillus plantarum NL42 .In order to remove some illegal restriction enzyme sites and add a HIS tag, the gene cwaA was synthesized by Shanghai Generay Biotech (Shanghai, China). The gene cut with restriction enzyme Hind III and NcoI was ligased with plasmid pNZ8148 cut with the same enzyme. Then the ligation product was transferred to E.coli and spread on plates containing 10 mg/L chloramphenicol. Colonies on the plates were randomly picked and inoculated in 1ml LB medium for 3 hours at 37℃, 200 rpm. 1 μl culture were added to the PCR system as template. As shown in Figure. 2, all the picked colonies had the brand of cwaA, illustrating that the plasmid pNZ-cwaA was successfully constructed.
Validation of autoaggregation of the recombinant L. lactis/pNZ-cwaA
To evaluate whether CwaA was involved in adhesion, we performed autoaggregation assays. The cells were collected and washed twice with o.1M PBS (pH=7.0).Then the cells were resuspended in 4ml PBS buffer with an initial OD600=0.5(A0) and the final OD600(At) of the upper most layer was measured after 4 hour cultivation in 30℃ without shaking.
-
Autoaggregation (%)=100-(At/A0)X100
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Sequence and Features