Difference between revisions of "Part:BBa K2812006"
(→Experimental Characterisation by TU-Eindhoven (2018)) |
(→Experimental Characterisation by TU-Eindhoven (2018)) |
||
| (3 intermediate revisions by 2 users not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K2812006 short</partinfo> | <partinfo>BBa_K2812006 short</partinfo> | ||
| − | Coding sequence for Pyocin S5 | + | Coding sequence for Pyocin S5 fused to the secretion signal peptide HlyA via a thrombin linker. TU-Eindhoven 2018 designed this part to secrete pyocin S5 from ''Escherichia coli'' to kill ''P. aeruginosa'' biofilms for the treatment of wound infections. For more information about our project, please visit our [http://2018.igem.org/Team:TU-Eindhoven wiki]. |
===Usage and Biology=== | ===Usage and Biology=== | ||
====Pyocin S5==== | ====Pyocin S5==== | ||
| − | + | Pyocin S5 is a bacteriocin produced by a specific strain of ''Pseudomonas aeruginosa''. Its lytic activity is initiated by binding of pyocin S5 to the outer membrane receptors of cells that are susceptible. Pyocin S5 consists out of three domains, a translocation, a receptor binding and a killing domain. Pyosin S5 induces membrane damage, resulting in the leakage of intracellular materials and subsequent cell death. It has been proven to have a bactericidal effect against seven ''P. aeruginosa''strains that are clinically relevant.<sup>1</sup> | |
====HlyA==== | ====HlyA==== | ||
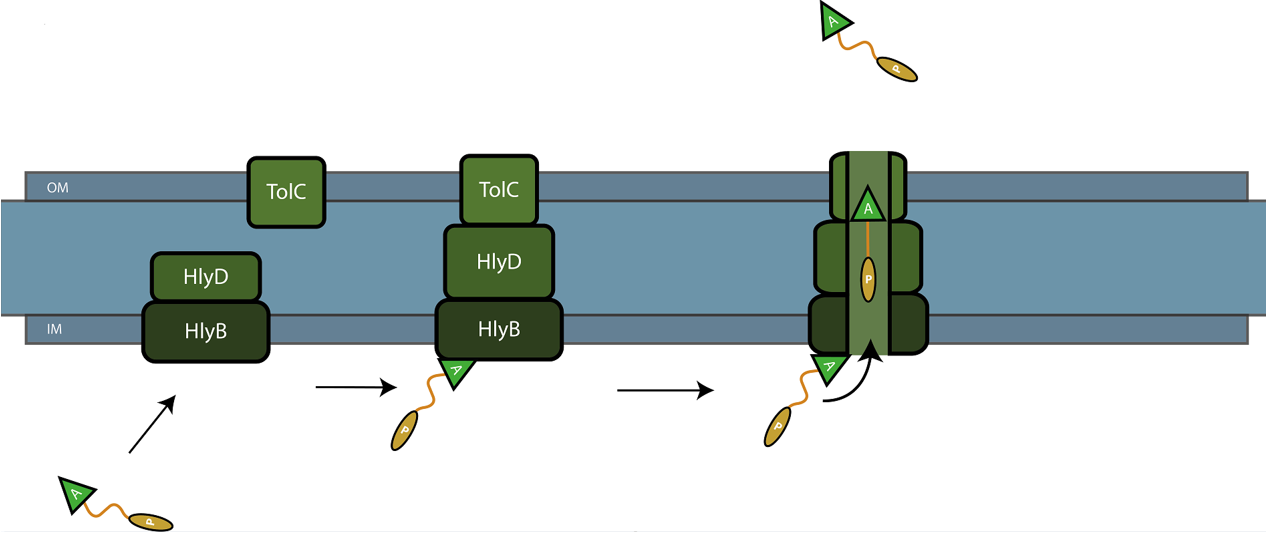
| − | The C-terminal sequence of Hemolysin A (residues 807-1024 of the ''E. coli'' HlyA gene) functions as a non-cleavable signal peptide for protein translocation via the Type I secretion pathway of Gram-negative bacteria. Here, HlyA is fused to the pyocin S5 for its secretion from ''E. coli'' BL21 (DE3) cells. In Type I secretion, single step transport of the target protein occurs from the cytoplasm to the extracellular environment. The HlyA is fused to the C-terminus of the target protein.<sup> | + | The C-terminal sequence of Hemolysin A (residues 807-1024 of the ''E. coli'' HlyA gene) functions as a non-cleavable signal peptide for protein translocation via the Type I secretion pathway of Gram-negative bacteria. Here, HlyA is fused to the pyocin S5 for its secretion from ''E. coli'' BL21 (DE3) cells. In Type I secretion, single step transport of the target protein occurs from the cytoplasm to the extracellular environment. The HlyA is fused to the C-terminus of the target protein.<sup>2</sup> It is a 23-kDa signal sequence that targets proteins for secretion via Type I secretion pathway. HlyA is secreted into the medium in a TolC and HlyB/D-dependent manner, in the presence of CaCl<sub>2</sub>.<sup>3</sup> It is recognized by the membrane translocation complex composed of HlyB and HlyD, which together with the TolC protein, will form a pore through the membrane.<sup>4</sup> This will lead to the secretion of HlyA-containing fusion proteins. Figure 1 illustrates the steps involved in the type I secretion of pyocin S5 as an HlyA fusion protein. |
====Thrombin linker & His-tag==== | ====Thrombin linker & His-tag==== | ||
| Line 22: | Line 22: | ||
[[File:BBa_K2812006_Gel_1kb_Pyosin.png|100px|right|Figure 1: 1% agarose gel of the colony PCR of the Pyocin S5-HlyA biobrick.]] | [[File:BBa_K2812006_Gel_1kb_Pyosin.png|100px|right|Figure 1: 1% agarose gel of the colony PCR of the Pyocin S5-HlyA biobrick.]] | ||
===Cloning=== | ===Cloning=== | ||
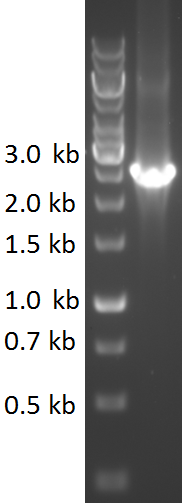
| − | TU-Eindhoven 2018 has characterized the biobrick <partinfo>BBa_K2812006</partinfo> at | + | TU-Eindhoven 2018 has characterized the biobrick <partinfo>BBa_K2812006</partinfo> at the DNA level. First, the pyocin S5-thrombin linker-HlyA-His-tag construct was synthesized by IDT and subsequently double digested and assembled into the digested linearized pSB1C3 backbone via ligation. The ligated construct was successfully transformed into ''E. coli'' NovaBlue, followed by a colony PCR using the VF2 and VR primers to investigate if the correct length has been inserted in the vector. The mixture was ran on a 1% agarose gel as can be seen in figure 1. The observed length of the brightest band corresponds with the expected length of 2531 basepairs, confirming that the desired construct has been succesfully ligated in pSB1C3 and subsequently transformed in ''E. coli'' NovaBlue. Next, the colonies with the correct insert were cultured in LB before a plasmid purification by a miniprep. The isolated plasmid DNA was sent for Sanger sequencing and the sequence could be confirmed. |
====Sources==== | ====Sources==== | ||
| − | 1) | + | 1) Ling, Hua, et al. A predicted S‐type pyocin shows a bactericidal activity against clinical Pseudomonas aeruginosa isolates through membrane damage. ''FEBS letters'' 584.15 (2010): 3354-3358. |
| − | 2) | + | 2) Thomas, S., Holland, I. B., & Schmitt, L. (2014). The Type 1 secretion pathway — The hemolysin system and beyond. ''Molecular Cell Research'', 1629-1641. |
| − | 3) | + | 3) Gray, L., Baker, K., Kenny, B., Mackman, N., Haigh, R., & Holland, I. (1989). A novel C-terminal signal sequence targets ''Escherichia coli'' haemolysin directly to the medium. ''J Cell Sci Suppl.'', 45-57. |
| − | 4 | + | 4) Wandersman, C., & Delepelaire, P. (1990 ). TolC, an ''Escherichia coli'' outer membrane protein required for hemolysin secretion. ''Proc Natl Acad Sci U S A.'', 4776-4780. |
| − | + | ||
| − | + | ||
<!-- --> | <!-- --> | ||
Latest revision as of 01:33, 17 October 2018
Coding sequence for Pyocin S5 with HlyA and His6-tag
Coding sequence for Pyocin S5 fused to the secretion signal peptide HlyA via a thrombin linker. TU-Eindhoven 2018 designed this part to secrete pyocin S5 from Escherichia coli to kill P. aeruginosa biofilms for the treatment of wound infections. For more information about our project, please visit our [http://2018.igem.org/Team:TU-Eindhoven wiki].
Usage and Biology
Pyocin S5
Pyocin S5 is a bacteriocin produced by a specific strain of Pseudomonas aeruginosa. Its lytic activity is initiated by binding of pyocin S5 to the outer membrane receptors of cells that are susceptible. Pyocin S5 consists out of three domains, a translocation, a receptor binding and a killing domain. Pyosin S5 induces membrane damage, resulting in the leakage of intracellular materials and subsequent cell death. It has been proven to have a bactericidal effect against seven P. aeruginosastrains that are clinically relevant.1
HlyA
The C-terminal sequence of Hemolysin A (residues 807-1024 of the E. coli HlyA gene) functions as a non-cleavable signal peptide for protein translocation via the Type I secretion pathway of Gram-negative bacteria. Here, HlyA is fused to the pyocin S5 for its secretion from E. coli BL21 (DE3) cells. In Type I secretion, single step transport of the target protein occurs from the cytoplasm to the extracellular environment. The HlyA is fused to the C-terminus of the target protein.2 It is a 23-kDa signal sequence that targets proteins for secretion via Type I secretion pathway. HlyA is secreted into the medium in a TolC and HlyB/D-dependent manner, in the presence of CaCl2.3 It is recognized by the membrane translocation complex composed of HlyB and HlyD, which together with the TolC protein, will form a pore through the membrane.4 This will lead to the secretion of HlyA-containing fusion proteins. Figure 1 illustrates the steps involved in the type I secretion of pyocin S5 as an HlyA fusion protein.
Thrombin linker & His-tag
The pyocin S5 and HlyA are linked via a thrombin linker. It is a short peptide sequence which can be cleaved by the enzyme thrombin, resulting in the removal of the HlyA domain. This avoids interference with the functionality of pyocin S5. C-terminal to the HlyA domain, a His-tag (6x repeated amino acid histidine) is attached. The His-tag can be used for detection of the fusion protein by e.g. Western Blot, but also for protein purification purposes as it facilitates binding to a nickel affinity column.
Fusion protein
The combination of all components described above, lead to a construct that can be (continuously) secreted by E. coli if the HlyB/D coding sequences have been co-transformed. Running the medium containing the fusion protein over a nickel affinity column and subsequent thrombin linker cleavage allows for the easy isolation of purified pyocin S5.
Experimental Characterisation by TU-Eindhoven (2018)
Cloning
TU-Eindhoven 2018 has characterized the biobrick BBa_K2812006 at the DNA level. First, the pyocin S5-thrombin linker-HlyA-His-tag construct was synthesized by IDT and subsequently double digested and assembled into the digested linearized pSB1C3 backbone via ligation. The ligated construct was successfully transformed into E. coli NovaBlue, followed by a colony PCR using the VF2 and VR primers to investigate if the correct length has been inserted in the vector. The mixture was ran on a 1% agarose gel as can be seen in figure 1. The observed length of the brightest band corresponds with the expected length of 2531 basepairs, confirming that the desired construct has been succesfully ligated in pSB1C3 and subsequently transformed in E. coli NovaBlue. Next, the colonies with the correct insert were cultured in LB before a plasmid purification by a miniprep. The isolated plasmid DNA was sent for Sanger sequencing and the sequence could be confirmed.
Sources
1) Ling, Hua, et al. A predicted S‐type pyocin shows a bactericidal activity against clinical Pseudomonas aeruginosa isolates through membrane damage. FEBS letters 584.15 (2010): 3354-3358.
2) Thomas, S., Holland, I. B., & Schmitt, L. (2014). The Type 1 secretion pathway — The hemolysin system and beyond. Molecular Cell Research, 1629-1641.
3) Gray, L., Baker, K., Kenny, B., Mackman, N., Haigh, R., & Holland, I. (1989). A novel C-terminal signal sequence targets Escherichia coli haemolysin directly to the medium. J Cell Sci Suppl., 45-57.
4) Wandersman, C., & Delepelaire, P. (1990 ). TolC, an Escherichia coli outer membrane protein required for hemolysin secretion. Proc Natl Acad Sci U S A., 4776-4780.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 137
Illegal BglII site found at 425
Illegal BglII site found at 2110 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 1200