Difference between revisions of "Part:BBa K2835003"
(→Ecotoxicity) |
Gomsemarii (Talk | contribs) |
||
| (22 intermediate revisions by 4 users not shown) | |||
| Line 2: | Line 2: | ||
<partinfo>BBa_K2835003 short</partinfo> | <partinfo>BBa_K2835003 short</partinfo> | ||
| − | This laccase from | + | This laccase from <i>Trametes versicolor</i> is originally a ligning degrading enzyme, but it is in fact capable of oxidizing a wide range of aromatic compounds. In our project, it has been used to inactivate the antibiotic sulfamethoxazole, one of the most persistent antibiotics found in the Baltic Sea [1]. |
| − | + | <br> | |
==Usage and Biology== | ==Usage and Biology== | ||
| − | Our BioBrick encodes a laccase from the fungus | + | Our BioBrick encodes a laccase from the fungus <i>Trametes versicolor</i> (PDB: 1GYC). Laccases are a class of multi-copper oxidases that fungi use to degrade lignin and be able to grow on wood [2]. Lignin is composed of a cluster of multiple phenolic groups, which are oxidized by laccases. With their natural affinity for phenolic-like structures, laccases have also been shown to degrade a large range of other phenolic compounds. For example, many reports show that laccases can be used to degrade pharmaceuticals - including estrogens, painkillers and antibiotics [3]. |
| − | BBa_K2835003 was created by modification of BBa_K500002 to remove the signal peptide sequence from the native host (first 60 bp) as annotated by UniProt (accession code: Q12718). Moreover, an N-terminal 6xHis-tag were added | + | BBa_K2835003 was created by modification of <partinfo>BBa_K500002</partinfo> to remove the signal peptide sequence from the native host (first 60 bp) as annotated by UniProt (accession code: Q12718). Moreover, an N-terminal 6xHis-tag were added. |
| − | + | ||
| − | + | ||
| + | In our project, we characterised this BioBrick by expressing it in the methylotrophic yeast <i>Pichia pastoris</i>. However, this BioBrick can be implemented in any host expression system, such as <i>S. cerevisiae</i> or <i>E. coli</i> by cloning it into an appropriate vector. | ||
| + | <br> | ||
====Expression==== | ====Expression==== | ||
| − | The BioBrick was cloned into the Invitrogen pPICZα A vector, which fuses it to the α-factor secretion signal from | + | The BioBrick was cloned into the Invitrogen pPICZα A vector, which fuses it to the α-factor secretion signal from <i>Saccharomyces cerevisiae</i>, and puts it under control of the methanol-inducible AOX1 promoter. After confirming the cloning by sequencing, the plasmid was electroporated into the X33 <i>Pichia pastoris</i> strain. The transformation was confirmed by colony PCR. |
Fourteen clones were picked to screen for enzyme production by cultivation in BMGY medium (containing glycerol) for 24h to gain biomass. Glycerol also derepresses the AOX1 promoter, which is repressed by glucose. Subsequently, the clones were cultivated in BMMY medium containing 1% methanol and 0.2 mM copper sulfate for 5 days. The addition of methanol induces protein production by activating the AOX1 promoter, whereas copper sulfate is required for proper folding of the laccase enzyme. | Fourteen clones were picked to screen for enzyme production by cultivation in BMGY medium (containing glycerol) for 24h to gain biomass. Glycerol also derepresses the AOX1 promoter, which is repressed by glucose. Subsequently, the clones were cultivated in BMMY medium containing 1% methanol and 0.2 mM copper sulfate for 5 days. The addition of methanol induces protein production by activating the AOX1 promoter, whereas copper sulfate is required for proper folding of the laccase enzyme. | ||
| − | Samples were taken and analyzed daily during cultivation in BMMY by performing an ABTS activity assay (100µl | + | Samples were taken and analyzed daily during cultivation in BMMY by performing an ABTS activity assay (100µl 2mM ABTS, 800µl 100mM citrate phosphate buffer pH 4.0, 100µl supernatant). On the second day of screening, we started observing a color change in the cuvettes after 10 to 30 minutes (Figure 1), versus no color change for the control supernatant (a blue color indicates formation of oxidized ABTS). On day 4 and 5, clear increases in absorbance were observed after 30 minutes. On the fifth day the increase in absorbance was even higher (Table 1). |
| − | [[Image:Blue cuvettes from ABTS Activity assay BBa K2835003.jpg|600px|thumb|center|<b>Figure 1 | + | [[Image:Blue cuvettes from ABTS Activity assay BBa K2835003.jpg|600px|thumb|center|<b>Figure 1.</b> ABTS assay cuvettes after 16 hours at room temperature, containing 100µl culture supernatant (clones 1-14, day 2), 100µl 2mM ABTS and 800µl citric acid-phosphate buffer pH 4.0. Cuvette labelled 31 contains only ABTS and buffer, cuvette labelled 32 contains only supernatant and buffer. Cuvette labelled C contains the wild-type control supernatant.]] |
| − | [[Image: | + | [[Image:Tablescreening2.png|500px|thumb|center|<b>Table 1.</b> Difference in absorbance measurements at 420nm at t=0 min vs t=30 min on day 4 and 5 for the top five laccase producing clones.]] |
| − | + | <br> | |
==Characterization== | ==Characterization== | ||
| − | Clone 2 was cultivated on a larger scale, with daily addition of 1% methanol and copper sulfate. After 5 days, the supernatant was collected by centrifugation and snap frozen in liquid nitrogen. A control culture, which was inoculated with the untransformed | + | Clone 2 was cultivated on a larger scale, with daily addition of 1% methanol and copper sulfate. After 5 days, the supernatant was collected by centrifugation and snap frozen in liquid nitrogen. A control culture, which was inoculated with the untransformed <i>Pichia pastoris</i> X33 strain was handled in the same way. |
====ABTS activity assay==== | ====ABTS activity assay==== | ||
| − | The | + | The BioBrick was characterised by measuring laccase activity in an ABTS assay. The assay was performed in a 96-well plate, adding 240 µL citrate phosphate buffer pH 4, 30 µL 2 mM ABTS (solved in ABTS buffer) and 30 µL of the culture supernatant. Supernatant of the clone 2 culture and the control culture were measured in triplicates. |
| − | As shown in Figure | + | As shown in Figure 2, the supernatant of clone 2 (expressing our BioBrick) showed a distinct activity compared to the control in the time frame of 70 min. |
| − | [[Image: | + | [[Image:Supernatantactivityassay1.png|600px|thumb|center|<b>Figure 2.</b> Oxidized ABTS product concentration over time at 30°C in 40 µL citrate phosphate buffer pH 4, 30 µL 2mM ABTS and 30 µL culture supernatant. Triplicates were performed for both clone 2 and the control. Error bars (grey) indicate the standard deviation (n=3). Statistically significant result is denoted with ****p≤0.0001 (comparing the mean of each group with a Mann-Whitney U test). ]] |
| − | We determined <i>K<sub>M</sub></i> and <i>v<sub>max</sub></i> for our wild type laccase by performing non-linear regression of a saturation curve on experimental data (Michaelis-Menten model). The kinetic parameters found with non-linear regression were: | + | We determined <i>K<sub>M</sub></i> and <i>v<sub>max</sub></i> for our wild-type laccase by performing non-linear regression of a saturation curve on experimental data (Michaelis-Menten model, Figure 3). The kinetic parameters found with non-linear regression were: |
<i>K<sub>M</sub></i> = 0.15 ± 0.0091 mg/ml ABTS | <i>K<sub>M</sub></i> = 0.15 ± 0.0091 mg/ml ABTS | ||
| Line 44: | Line 44: | ||
<i>v<sub>max</sub></i> = 29 ± 0.84 μM/min | <i>v<sub>max</sub></i> = 29 ± 0.84 μM/min | ||
| − | [[Image:iGEM Stockholm 2018 MM-model.jpg|400px|thumb|center|<b>Figure | + | [[Image:iGEM Stockholm 2018 MM-model.jpg|400px|thumb|center|<b>Figure 3.</b> A comparison of the enzymatic activity of wild-type laccase in different substrate concentrations at pH 4.0, 30 °C. Enzyme concentration was approximately 0.0069 mg/ml in the reactions. Error bars indicate standard deviation (n=3). The blue line represents the Michaelis-Menten model obtained from non-linear regression (R2 = 0.9952).]] |
| + | |||
===Ecotoxicity=== | ===Ecotoxicity=== | ||
| − | |||
| − | [[Image:IGEM_Stockholm_2018_EC50_table.png|500px|center|]] | + | We were able to successfully measure the EC50 values for both the transformation reactions with our wild type laccase and pure SMX. We tested transformation products (TPs) generated from two different concentrations of SMX and wild type laccase; a low concentration of 5 mg/mL and a high concentration of 7.6 mg/mL SMX. Using the bioluminescent bacteria <i>A. fischeri</i>, we were able to measure light inhibition which correlates with <i>A. fischeri</i> death. SMX is toxic for <i>A. fischeri</i> which is shown since the EC50 value was approximately 50 mg/L for pure SMX, for both 5 mg/mL and 7.6 mg/mL of SMX (Table 2). For the TPs we observed a 3-fold increase of the EC50. This shows that the wild type laccase is degrading SMX, and the products of the reaction are not toxic for the environment. |
| + | |||
| + | |||
| + | [[Image:IGEM_Stockholm_2018_EC50_table.png|500px|thumb|center|<b>Table 2.</b> EC50 values found for pure SMX and transformation products after SMX degradation by laccase.]] | ||
==References== | ==References== | ||
| + | [1] Helcom (2017). Pharmaceuticals in the aquatic environment of the Baltic Sea region – A status report. Emerging pollutants in water. [online] Paris 07 SP, France: The United Nations Educational, Scientific and Cultural Organization and Helcom. | ||
| + | [2] Solomon E., Sundaram U., Machonkin T. Multicopper Oxidases and Oxygenases. Chem Rev. 1996;96:2563–2606. | ||
| + | |||
| + | [3] Naghdi, M., Taheran, M., Satinder, K., Kermanshahi-pour, A., Verma, M. and Surampalli, R.Y. Removal of pharmaceutical compounds in water and wastewater using fungal oxidoreductase enzymes. Environmental Pollution vol 234, March 2018, pages 190-213. | ||
| + | ===iGEM2020-KSA_KOREA=== | ||
| + | <p style='margin-right: 0px; margin-bottom: 2px; margin-left: 0px; font-stretch: normal; font-size: 14px; line-height: normal; font-family: "Helvetica Neue";'><strong>Laccase from <em>Trametes versicolor</em></strong></p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'>The Laccase from <em>Trametes versicolor</em> is a class of multi-copper oxidase, which is one of the most components of ligninolytic complex. Laccase is capable of oxidizing the aromatic compounds, including moieties typically found in lignin. In the project, our purpose is to show the possibility of producing high quality and eco-friendly recycled paper with low lignin content, which used Laccase in the process of recycled paper production process.</p><p style='margin-right: 0px; margin-bottom: 2px; margin-left: 0px; font-stretch: normal; font-size: 14px; line-height: normal; font-family: "Helvetica Neue";'><strong>Usage and Biology</strong></p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'>Our BioBrick (BBa_K3654000) encodes a Laccase from the fungus <em>Trametes versicolor</em>, which was reported to have the highest redox potential among Laccases. Laccases are the most important components of ligninolytic enzyme of wood-destroying microorganisms. Lignin is one of the most abundant naturally cross-linked phenolic biopolymers, which combine cellulose and comprise 20~35% of the dry weight of plant cell wall. However, lignin exerts negative impact on the utilization of plant biomass in pulp and paper industry. The presence of lignin in paper pulp reduces paper permanence and discolors it over time. Recently, several works are done to search specific mechanisms of enzymatic lignin oxidation that laccase perform, presenting a good potential for the base of new environmentally friendly technologies in pulp and paper industry. Since Laccase has the ability to oxidize various substrates, it is applied to multiple fields, including textile dye bleaching and biomediation.</p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'>BBa_K3654000 was created by codon optimization for <em>Pichia</em> <em>pastoris</em> from BBa_K500002. It was cloned into the Invitrogen pPinkα-HC vector, which fuses it to the α-factor secretion signal from <em>Saccharomyces cerevisiae</em>, and puts it under the control of the methanol-inducible AOX1 promoter. In our project, this BioBrick was characterized by expressing in the methylotrophic yeast <em>P. pastoris</em> strain PichiaPink.</p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'><strong>Selection</strong></p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'>The BioBlock was cloned into the <em>P. pastoris</em> expression vector pPinkα-HC (Invitrogen), which has the α-mating factor secretion signal sequence, and methanol-inducible AOX1promoter. After cloning and confirming DNA sequence, the plasmid was transformed into the methylotrophic yeast <em>P. pastoris</em> strain PichiaPink by electrophoration. Recombinant Laccase expressing transformants were selected with minimal methanol (MM) plates containing 0.3mM CuSO4 and 0.2mM ABTS (Fig. 1).</p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'>https://2020.igem.org/wiki/images/3/3e/T--KSA_KOREA--fig-1.png</p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'><strong>Fig. 1.</strong> Selection of recombinant Laccase expressing transformants using MM plates containing 0.3mM CuSO4and 0.2 mM ABTS.</p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'><strong>Expression</strong></p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'>Recombinant Laccase expressing transformant was grown in BMGY medium (containing 1% glycerol) for 2 days. Glycerol as an energy source was used to prevent the possibility of catabolite repression by trace amounts of glucose. The culture medium for expression was BMMY medium containing 0.5 % methanol and 0.3 mM copper sulfate (CuSO4). Methanol was added daily to a final concentration of 0.5 % to induce protein production by activating the methanol-inducible AOX1promoter for 13 days.</p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue"; min-height: 14px;'><br></p><p style='margin-right: 0px; margin-bottom: 2px; margin-left: 0px; font-stretch: normal; font-size: 14px; line-height: normal; font-family: "Helvetica Neue";'><strong>Characterization</strong></p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'>Recombinant Laccase expressing transformant was grown in BMMY medium containing 0.5 % methanol and 0.3 mM CuSO4 for 13 days. Every day, samples were taken 1 ml from the culture and were precipitated by adding ammonium sulfate to a final mass concentration of 80%. The concentrated proteins were dissolved in 100 μl of 100 mM sodium acetate buffer (pH 5.0). Recombinant Laccase production was analyzed by Coomassie-stained SDS-PAGE and enzyme activity with ABTS assay. A control culture was inoculated with the untransformed <em>Pichia pastoris</em> strain PichiaPink, and was handled in the same way.</p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'><strong>SDS-Polyacrylamide Gel Electrophoresis</strong></p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'>The expression pattern of BioBrick was characterized by SDS-PAGE analysis. SDS-PAGE with 10 % separating gel and 4 % stacking gel was performed as described by Laemmli. Protein bands were stained with Coomassie blue.</p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue"; min-height: 14px;'><br></p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'>https://2020.igem.org/wiki/images/9/9b/T--KSA_KOREA--fig-2.png</p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'><strong>Fig. 2.</strong> Coomassie staining of SDS-PAGE gel on the accumulation of recombinant Laccase in the supernatant of samples taken at each time points.</p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue"; min-height: 14px;'><br></p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'>The expected sizes of recombinant Laccase were about 55.8KDa without glycosylation. However, since Laccase is a known glycosylated protein, it is likely that recombinant Laccase would shift to a higher size. As such, the observed size of recombinant Laccase was about over 70KDa (Fig. 2).</p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue"; min-height: 14px;'><br></p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue"; min-height: 14px;'><br></p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'><strong>Enzyme Activity</strong></p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'>The enzyme activity of BioBrick was characterized by ABTS analysis (Fig. 3). Recombinant Laccase activity assays contained 100 mM sodium acetate (pH5.0), 0.5 mM ABTS, and 100 μl of culture supernatant in a total volume of 1 ml. Recombinant Laccase activity was measured as the increase in absorbance at 420 nm and was expressed as activity units per liter. One unit is 1 μmol ABTS oxidized per minute. The molar extinction coefficient for ABTS at 420 nm was taken to be 36,000M-1cm-1.</p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'>https://2020.igem.org/wiki/images/4/49/T--KSA_KOREA--fig-3.png</p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'><strong>Fig. 3.</strong> Recombinant Laccase production by <em>Pichia pastoris</em> transformants in buffered minimal media. (A) Time course of color change in ABTS assay due to recombinant Laccase activity. (B) Time course of recombinant Laccase activity during a representative cultivation. Triplicates were performed for determination of recombinant Laccase activity. Error bars indicate the standard deviation (n=3).</p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'>Recombinant Laccase production by <em>P. pastoris</em> in BMMY liquid medium was monitored for 13 days in the shaking flask. As shown in Fig. 3, maximum Laccase activity (7.67 U/L) was reached on day 9. Therefore, we determined the optimal culture period as 9 days.</p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue"; min-height: 14px;'><br></p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'><strong>Lignin Degradation Activity</strong></p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'>The lignin degradation activity of BioBrick was measured by Klason method. 2 U recombinant Laccase was added to 0.5 g of each waste papers, and incubated with 10 mM 1-hydroxybenzotriazole (HBT) at 26 ℃ for 24 hrs. Total lignin degradation of each waste papers by recombinant Laccase was determined using Klason method (Fig. 4).</p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'>https://2020.igem.org/wiki/images/0/07/T--KSA_KOREA--fig-4.png</p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; text-align: justify; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'><strong>Fig. 4. </strong>Total lignin degradation of different waste papers by recombinant Laccase with 10 mM HBT for 24 hrs at 26 ℃. Average degradation rate of total lignin (%) was 35.02%, and the degradation rate was similar regardless of the type of paper.</p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'>Recombinant Laccase with 10 mM HBT degraded an average of 35.02% of lignin in four types of wastepaper, and there was no significantly difference according to the type of waste paper (Fig. 4). This result shows the possibility of producing high quality recycled paper with low lignin content using recombinant Laccase, which was produced by our BioBrick.</p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue"; min-height: 14px;'><br></p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; text-align: justify; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'><strong>Paper Permanence fImprovement </strong></p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'>We made recycled paper from wastepaper using lignin degradation activity of BioBrick (Fig. 5). To make recycled papers, 5 U recombinant Laccase was added to 5 g of each waste papers, and incubated with 10 mM 1-hydroxybenzotriazole (HBT) at 26 ℃ for 24 hrs. Then recycled papers were compared for the degree of paper color change by sunlight irradiation with control (Fig. 6).</p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; text-align: justify; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue"; min-height: 14px;'><br></p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; text-align: justify; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue"; min-height: 14px;'><br></p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'>https://2020.igem.org/wiki/images/b/bd/T--KSA_KOREA--fig-5.png</p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'><strong>Fig. 5.</strong> Comparison between recycled papers made from pulp treated with recombinant laccase and untreated recycled papers. Lignin of wastepaper was degraded by recombinant Laccase with 10 mM HBT for 24 hrs at 26 ℃.</p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'>Recycled papers made with pulp treated with laccase were brighter than recycled papers made with untreated pulp. However, treated recycled papers were yellower than untreated recycled paper. The yellow color of treated recycled paper is presumably due to staining by recombinant Laccase. Treated recycled papers had improved texture compared to untreated recycled papers. This improved texture of recycled papers was presumably due to the relative increase of cellulose content caused by laccase degrading lignin.</p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'>https://2020.igem.org/wiki/images/7/76/T--KSA_KOREA--fig-6.png</p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'><strong>Fig. 6.</strong> Comparison of aging effect between recycled papers treated with recombinant laccase and untreated recycled papers. Categorized by pulp source: Newspaper and box paper</p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue"; min-height: 14px;'><br></p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'>Lignin removal is essential to producing high quality paper, as lignin in paper <span style="text-decoration: underline;">pulp</span> that reduces paper permanence and contributes to the yellowing of paper over time. Therefore, we compared the yellowing effect by ultraviolet irradiation between recycled papers and waste papers. Recycled papers that were treated with recombinant Laccase exhibited less color shift compared to untreated recycled papers. This result shows that degrading lignin with recombinant laccase results in visually observable increase in paper quality. If these experimental results are further improved and applied to mass production of high-quality recycled paper from wastepaper using recombinant Laccase, it could contribute to the protection of ecosystem and environment.</p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue"; min-height: 14px;'><br></p><p style='margin-right: 0px; margin-bottom: 2px; margin-left: 0px; font-stretch: normal; font-size: 14px; line-height: normal; font-family: "Helvetica Neue";'><strong>Reference</strong></p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; text-align: justify; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'>[1] Kurniawati S. and Nicell J.A. (2008) Characterization of <em>Trametes</em> <em>versicolor</em> laccase for the transformation of aqueous phenol. Bioresour. Technol. 99: 7825–7834.</p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; text-align: justify; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'>[2] Faravelli T., Frassoldati, A., Migliavacca, G., Ranzi, E. (2010) Detailed kinetic modeling of the thermal degradation of lignins. Biomass & Bioenergy. 34(3): 290-301.</p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; text-align: justify; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'>[3] Verma, S.R. and Dwivedi, U.N. (2014) Lignin genetic engineering for improvement of wood quality: Applications in paper and textile industries, fodder and bioenergy production, South African Journal of Botany. 91: 107-125.</p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; text-align: justify; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'>[4] Litwińska K., Bischoff F., Matthes F., Bode R., Rutten T., Kunze G. (2019) Characterization of recombinant laccase from Trametes versicolor synthesized by Arxula adeninivorans and its application in the degradation of pharmaceuticals. AMB Express. 9(1):102. doi: 10.1186/s13568-019-0832-3.</p><p style='margin-right: 0px; margin-bottom: 0px; margin-left: 0px; text-align: justify; font-stretch: normal; font-size: 12px; line-height: normal; font-family: "Helvetica Neue";'>[5] Laemmli, U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227:680-685.</p><p><br></p> | ||
<!-- --> | <!-- --> | ||
Latest revision as of 23:04, 27 October 2020
His-tagged laccase from Trametes versicolor
This laccase from Trametes versicolor is originally a ligning degrading enzyme, but it is in fact capable of oxidizing a wide range of aromatic compounds. In our project, it has been used to inactivate the antibiotic sulfamethoxazole, one of the most persistent antibiotics found in the Baltic Sea [1].
Usage and Biology
Our BioBrick encodes a laccase from the fungus Trametes versicolor (PDB: 1GYC). Laccases are a class of multi-copper oxidases that fungi use to degrade lignin and be able to grow on wood [2]. Lignin is composed of a cluster of multiple phenolic groups, which are oxidized by laccases. With their natural affinity for phenolic-like structures, laccases have also been shown to degrade a large range of other phenolic compounds. For example, many reports show that laccases can be used to degrade pharmaceuticals - including estrogens, painkillers and antibiotics [3].
BBa_K2835003 was created by modification of BBa_K500002 to remove the signal peptide sequence from the native host (first 60 bp) as annotated by UniProt (accession code: Q12718). Moreover, an N-terminal 6xHis-tag were added.
In our project, we characterised this BioBrick by expressing it in the methylotrophic yeast Pichia pastoris. However, this BioBrick can be implemented in any host expression system, such as S. cerevisiae or E. coli by cloning it into an appropriate vector.
Expression
The BioBrick was cloned into the Invitrogen pPICZα A vector, which fuses it to the α-factor secretion signal from Saccharomyces cerevisiae, and puts it under control of the methanol-inducible AOX1 promoter. After confirming the cloning by sequencing, the plasmid was electroporated into the X33 Pichia pastoris strain. The transformation was confirmed by colony PCR.
Fourteen clones were picked to screen for enzyme production by cultivation in BMGY medium (containing glycerol) for 24h to gain biomass. Glycerol also derepresses the AOX1 promoter, which is repressed by glucose. Subsequently, the clones were cultivated in BMMY medium containing 1% methanol and 0.2 mM copper sulfate for 5 days. The addition of methanol induces protein production by activating the AOX1 promoter, whereas copper sulfate is required for proper folding of the laccase enzyme.
Samples were taken and analyzed daily during cultivation in BMMY by performing an ABTS activity assay (100µl 2mM ABTS, 800µl 100mM citrate phosphate buffer pH 4.0, 100µl supernatant). On the second day of screening, we started observing a color change in the cuvettes after 10 to 30 minutes (Figure 1), versus no color change for the control supernatant (a blue color indicates formation of oxidized ABTS). On day 4 and 5, clear increases in absorbance were observed after 30 minutes. On the fifth day the increase in absorbance was even higher (Table 1).

Characterization
Clone 2 was cultivated on a larger scale, with daily addition of 1% methanol and copper sulfate. After 5 days, the supernatant was collected by centrifugation and snap frozen in liquid nitrogen. A control culture, which was inoculated with the untransformed Pichia pastoris X33 strain was handled in the same way.
ABTS activity assay
The BioBrick was characterised by measuring laccase activity in an ABTS assay. The assay was performed in a 96-well plate, adding 240 µL citrate phosphate buffer pH 4, 30 µL 2 mM ABTS (solved in ABTS buffer) and 30 µL of the culture supernatant. Supernatant of the clone 2 culture and the control culture were measured in triplicates.
As shown in Figure 2, the supernatant of clone 2 (expressing our BioBrick) showed a distinct activity compared to the control in the time frame of 70 min.

We determined KM and vmax for our wild-type laccase by performing non-linear regression of a saturation curve on experimental data (Michaelis-Menten model, Figure 3). The kinetic parameters found with non-linear regression were:
KM = 0.15 ± 0.0091 mg/ml ABTS
vmax = 29 ± 0.84 μM/min

Ecotoxicity
We were able to successfully measure the EC50 values for both the transformation reactions with our wild type laccase and pure SMX. We tested transformation products (TPs) generated from two different concentrations of SMX and wild type laccase; a low concentration of 5 mg/mL and a high concentration of 7.6 mg/mL SMX. Using the bioluminescent bacteria A. fischeri, we were able to measure light inhibition which correlates with A. fischeri death. SMX is toxic for A. fischeri which is shown since the EC50 value was approximately 50 mg/L for pure SMX, for both 5 mg/mL and 7.6 mg/mL of SMX (Table 2). For the TPs we observed a 3-fold increase of the EC50. This shows that the wild type laccase is degrading SMX, and the products of the reaction are not toxic for the environment.
References
[1] Helcom (2017). Pharmaceuticals in the aquatic environment of the Baltic Sea region – A status report. Emerging pollutants in water. [online] Paris 07 SP, France: The United Nations Educational, Scientific and Cultural Organization and Helcom.
[2] Solomon E., Sundaram U., Machonkin T. Multicopper Oxidases and Oxygenases. Chem Rev. 1996;96:2563–2606.
[3] Naghdi, M., Taheran, M., Satinder, K., Kermanshahi-pour, A., Verma, M. and Surampalli, R.Y. Removal of pharmaceutical compounds in water and wastewater using fungal oxidoreductase enzymes. Environmental Pollution vol 234, March 2018, pages 190-213.
iGEM2020-KSA_KOREA
Laccase from Trametes versicolor
The Laccase from Trametes versicolor is a class of multi-copper oxidase, which is one of the most components of ligninolytic complex. Laccase is capable of oxidizing the aromatic compounds, including moieties typically found in lignin. In the project, our purpose is to show the possibility of producing high quality and eco-friendly recycled paper with low lignin content, which used Laccase in the process of recycled paper production process.
Usage and Biology
Our BioBrick (BBa_K3654000) encodes a Laccase from the fungus Trametes versicolor, which was reported to have the highest redox potential among Laccases. Laccases are the most important components of ligninolytic enzyme of wood-destroying microorganisms. Lignin is one of the most abundant naturally cross-linked phenolic biopolymers, which combine cellulose and comprise 20~35% of the dry weight of plant cell wall. However, lignin exerts negative impact on the utilization of plant biomass in pulp and paper industry. The presence of lignin in paper pulp reduces paper permanence and discolors it over time. Recently, several works are done to search specific mechanisms of enzymatic lignin oxidation that laccase perform, presenting a good potential for the base of new environmentally friendly technologies in pulp and paper industry. Since Laccase has the ability to oxidize various substrates, it is applied to multiple fields, including textile dye bleaching and biomediation.
BBa_K3654000 was created by codon optimization for Pichia pastoris from BBa_K500002. It was cloned into the Invitrogen pPinkα-HC vector, which fuses it to the α-factor secretion signal from Saccharomyces cerevisiae, and puts it under the control of the methanol-inducible AOX1 promoter. In our project, this BioBrick was characterized by expressing in the methylotrophic yeast P. pastoris strain PichiaPink.
Selection
The BioBlock was cloned into the P. pastoris expression vector pPinkα-HC (Invitrogen), which has the α-mating factor secretion signal sequence, and methanol-inducible AOX1promoter. After cloning and confirming DNA sequence, the plasmid was transformed into the methylotrophic yeast P. pastoris strain PichiaPink by electrophoration. Recombinant Laccase expressing transformants were selected with minimal methanol (MM) plates containing 0.3mM CuSO4 and 0.2mM ABTS (Fig. 1).
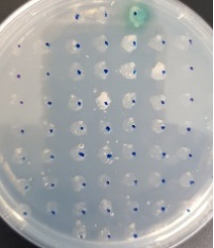
Fig. 1. Selection of recombinant Laccase expressing transformants using MM plates containing 0.3mM CuSO4and 0.2 mM ABTS.
Expression
Recombinant Laccase expressing transformant was grown in BMGY medium (containing 1% glycerol) for 2 days. Glycerol as an energy source was used to prevent the possibility of catabolite repression by trace amounts of glucose. The culture medium for expression was BMMY medium containing 0.5 % methanol and 0.3 mM copper sulfate (CuSO4). Methanol was added daily to a final concentration of 0.5 % to induce protein production by activating the methanol-inducible AOX1promoter for 13 days.
Characterization
Recombinant Laccase expressing transformant was grown in BMMY medium containing 0.5 % methanol and 0.3 mM CuSO4 for 13 days. Every day, samples were taken 1 ml from the culture and were precipitated by adding ammonium sulfate to a final mass concentration of 80%. The concentrated proteins were dissolved in 100 μl of 100 mM sodium acetate buffer (pH 5.0). Recombinant Laccase production was analyzed by Coomassie-stained SDS-PAGE and enzyme activity with ABTS assay. A control culture was inoculated with the untransformed Pichia pastoris strain PichiaPink, and was handled in the same way.
SDS-Polyacrylamide Gel Electrophoresis
The expression pattern of BioBrick was characterized by SDS-PAGE analysis. SDS-PAGE with 10 % separating gel and 4 % stacking gel was performed as described by Laemmli. Protein bands were stained with Coomassie blue.
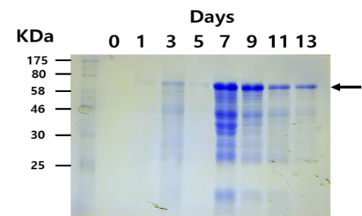
Fig. 2. Coomassie staining of SDS-PAGE gel on the accumulation of recombinant Laccase in the supernatant of samples taken at each time points.
The expected sizes of recombinant Laccase were about 55.8KDa without glycosylation. However, since Laccase is a known glycosylated protein, it is likely that recombinant Laccase would shift to a higher size. As such, the observed size of recombinant Laccase was about over 70KDa (Fig. 2).
Enzyme Activity
The enzyme activity of BioBrick was characterized by ABTS analysis (Fig. 3). Recombinant Laccase activity assays contained 100 mM sodium acetate (pH5.0), 0.5 mM ABTS, and 100 μl of culture supernatant in a total volume of 1 ml. Recombinant Laccase activity was measured as the increase in absorbance at 420 nm and was expressed as activity units per liter. One unit is 1 μmol ABTS oxidized per minute. The molar extinction coefficient for ABTS at 420 nm was taken to be 36,000M-1cm-1.
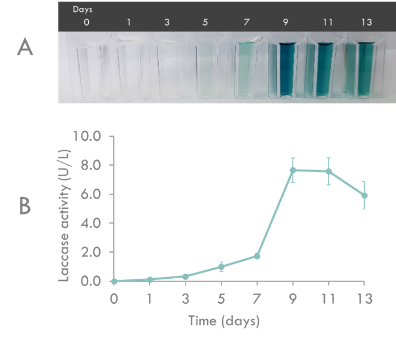
Fig. 3. Recombinant Laccase production by Pichia pastoris transformants in buffered minimal media. (A) Time course of color change in ABTS assay due to recombinant Laccase activity. (B) Time course of recombinant Laccase activity during a representative cultivation. Triplicates were performed for determination of recombinant Laccase activity. Error bars indicate the standard deviation (n=3).
Recombinant Laccase production by P. pastoris in BMMY liquid medium was monitored for 13 days in the shaking flask. As shown in Fig. 3, maximum Laccase activity (7.67 U/L) was reached on day 9. Therefore, we determined the optimal culture period as 9 days.
Lignin Degradation Activity
The lignin degradation activity of BioBrick was measured by Klason method. 2 U recombinant Laccase was added to 0.5 g of each waste papers, and incubated with 10 mM 1-hydroxybenzotriazole (HBT) at 26 ℃ for 24 hrs. Total lignin degradation of each waste papers by recombinant Laccase was determined using Klason method (Fig. 4).
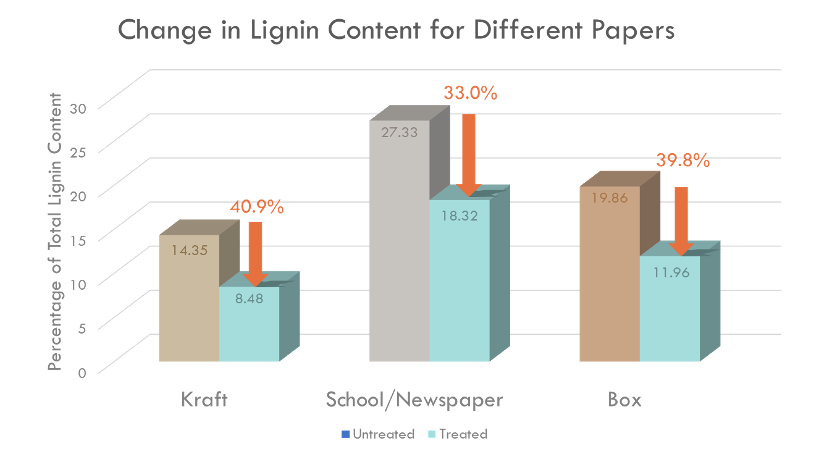
Fig. 4. Total lignin degradation of different waste papers by recombinant Laccase with 10 mM HBT for 24 hrs at 26 ℃. Average degradation rate of total lignin (%) was 35.02%, and the degradation rate was similar regardless of the type of paper.
Recombinant Laccase with 10 mM HBT degraded an average of 35.02% of lignin in four types of wastepaper, and there was no significantly difference according to the type of waste paper (Fig. 4). This result shows the possibility of producing high quality recycled paper with low lignin content using recombinant Laccase, which was produced by our BioBrick.
Paper Permanence fImprovement
We made recycled paper from wastepaper using lignin degradation activity of BioBrick (Fig. 5). To make recycled papers, 5 U recombinant Laccase was added to 5 g of each waste papers, and incubated with 10 mM 1-hydroxybenzotriazole (HBT) at 26 ℃ for 24 hrs. Then recycled papers were compared for the degree of paper color change by sunlight irradiation with control (Fig. 6).

Fig. 5. Comparison between recycled papers made from pulp treated with recombinant laccase and untreated recycled papers. Lignin of wastepaper was degraded by recombinant Laccase with 10 mM HBT for 24 hrs at 26 ℃.
Recycled papers made with pulp treated with laccase were brighter than recycled papers made with untreated pulp. However, treated recycled papers were yellower than untreated recycled paper. The yellow color of treated recycled paper is presumably due to staining by recombinant Laccase. Treated recycled papers had improved texture compared to untreated recycled papers. This improved texture of recycled papers was presumably due to the relative increase of cellulose content caused by laccase degrading lignin.

Fig. 6. Comparison of aging effect between recycled papers treated with recombinant laccase and untreated recycled papers. Categorized by pulp source: Newspaper and box paper
Lignin removal is essential to producing high quality paper, as lignin in paper pulp that reduces paper permanence and contributes to the yellowing of paper over time. Therefore, we compared the yellowing effect by ultraviolet irradiation between recycled papers and waste papers. Recycled papers that were treated with recombinant Laccase exhibited less color shift compared to untreated recycled papers. This result shows that degrading lignin with recombinant laccase results in visually observable increase in paper quality. If these experimental results are further improved and applied to mass production of high-quality recycled paper from wastepaper using recombinant Laccase, it could contribute to the protection of ecosystem and environment.
Reference
[1] Kurniawati S. and Nicell J.A. (2008) Characterization of Trametes versicolor laccase for the transformation of aqueous phenol. Bioresour. Technol. 99: 7825–7834.
[2] Faravelli T., Frassoldati, A., Migliavacca, G., Ranzi, E. (2010) Detailed kinetic modeling of the thermal degradation of lignins. Biomass & Bioenergy. 34(3): 290-301.
[3] Verma, S.R. and Dwivedi, U.N. (2014) Lignin genetic engineering for improvement of wood quality: Applications in paper and textile industries, fodder and bioenergy production, South African Journal of Botany. 91: 107-125.
[4] Litwińska K., Bischoff F., Matthes F., Bode R., Rutten T., Kunze G. (2019) Characterization of recombinant laccase from Trametes versicolor synthesized by Arxula adeninivorans and its application in the degradation of pharmaceuticals. AMB Express. 9(1):102. doi: 10.1186/s13568-019-0832-3.
[5] Laemmli, U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227:680-685.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 544
Illegal BglII site found at 1240 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]


