Difference between revisions of "Part:BBa K2812004"
(→Experimental Characterisation by TU Eindhoven (2018)) |
(→Functional improvement of BBa_K748002) |
||
| (26 intermediate revisions by 2 users not shown) | |||
| Line 7: | Line 7: | ||
===Usage & Biology=== | ===Usage & Biology=== | ||
====Lysostaphin==== | ====Lysostaphin==== | ||
| − | Lysostaphin is an antimicrobial agent produced by ''Staphylococcus simulans''. | + | Lysostaphin is an antimicrobial agent produced by ''Staphylococcus simulans''. It targets the cell wall peptidoglycan found in certain ''Staphylococci'' by cleaving its cross-linking pentaglycine bridges. Among others, it is effective for degrading ''Staphylococcus aureus'' biofilms.<sup>1</sup> The encoding part of the lysostaphin has been derived from <partinfo>BBa_K748002</partinfo>, which was made by iGEM Harbin 2012 and was also used by iGEM Stockholm 2016. iGEM Eindhoven 2018 codon optimized this lysostaphin construct. Lysostaphin belongs to the major class of antimicrobial proteins and peptides known as bacteriocins. Bacteriocins are proteins or peptides produced by bacteria, displaying a bactericidal activity against other subpopulations of bacteria.<sup>2</sup> The cell wall degradation capability of lysostaphin derives from its endopeptidase activity on pentaglycine cross-bridges in the peptidoglycan layer. Specific cleavage between the third and fourth glycine residue leads to the destruction of the peptidoglycan layer and subsequent lysis of the bacteria. |
| − | targets the cell wall peptidoglycan found in certain Staphylococci by cleaving its cross-linking pentaglycine bridges. Among others, it is effective for degrading | + | |
====HlyA==== | ====HlyA==== | ||
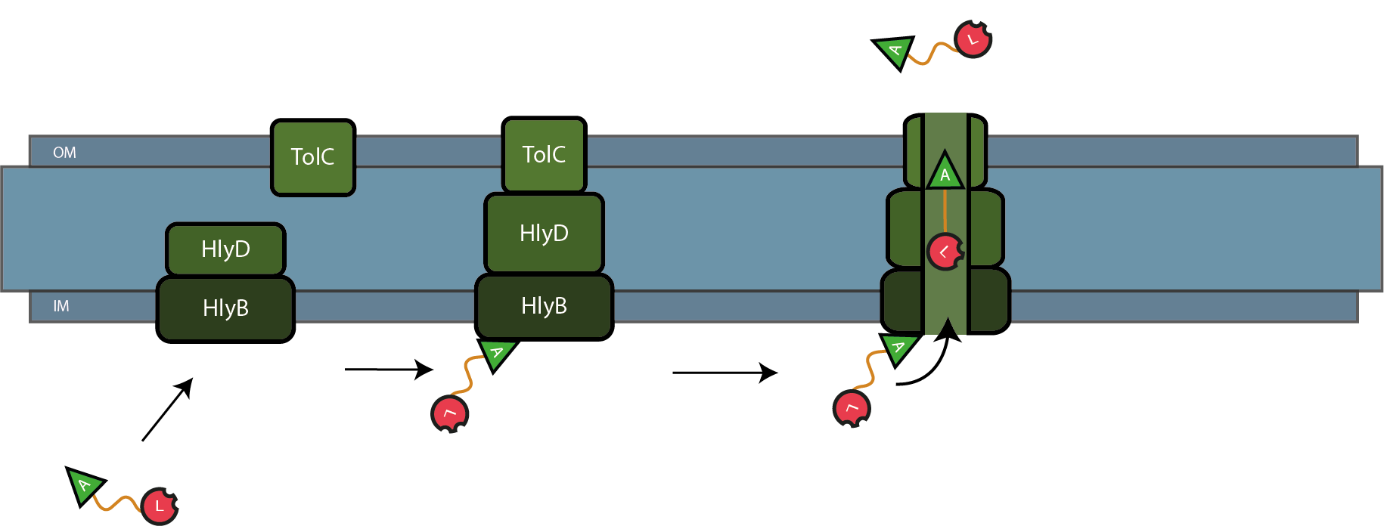
| − | The C-terminal sequence of Hemolysin A (residues 807-1024 of the ''E. coli'' HlyA gene) functions as a non-cleavable signal peptide for protein translocation via the Type I secretion pathway of Gram-negative bacteria. Here, HlyA is fused to the lysostaphin for its secretion from ''E. coli'' BL21 (DE3) cells. In Type I secretion, single step transport of the target protein occurs from the cytoplasm to the extracellular environment. The HlyA is fused to the C-terminus of the target protein.<sup>3</sup> It is a 23-kDa signal sequence that targets proteins for secretion via Type I secretion pathway. HlyA is secreted into the medium in a TolC and HlyB/D-dependent manner.<sup>4</sup> It is recognized by the membrane translocation complex composed of HlyB and HlyD, which together with the TolC protein, will form a pore through the membrane.<sup>5</sup> This will lead to the secretion of HlyA-containing fusion proteins. Figure 1 illustrates the steps involved in the type I secretion of lysostaphin as an HlyA fusion protein. | + | The C-terminal sequence of Hemolysin A (residues 807-1024 of the ''E. coli'' HlyA gene) functions as a non-cleavable signal peptide for protein translocation via the Type I secretion pathway of Gram-negative bacteria. Here, HlyA is fused to the lysostaphin for its secretion from ''E. coli'' BL21 (DE3) cells. In Type I secretion, single step transport of the target protein occurs from the cytoplasm to the extracellular environment. The HlyA is fused to the C-terminus of the target protein.<sup>3</sup> It is a 23-kDa signal sequence that targets proteins for secretion via Type I secretion pathway. HlyA is secreted into the medium in a TolC and HlyB/D-dependent manner, in the presence of CaCl<sub>2</sub>.<sup>4</sup> It is recognized by the membrane translocation complex composed of HlyB and HlyD, which together with the TolC protein, will form a pore through the membrane.<sup>5</sup> This will lead to the secretion of HlyA-containing fusion proteins. Figure 1 illustrates the steps involved in the type I secretion of lysostaphin as an HlyA fusion protein. |
====Thrombin linker & His-tag==== | ====Thrombin linker & His-tag==== | ||
| Line 20: | Line 19: | ||
[[File:BBa_K2812004_secrety_lyso.png|900px|thumb|centre|Figure 1: Type I secretion of lysostaphin via HlyA, HlyB, HlyD and TolC. IM = inner membrane; OM = outer membrane; A = HlyA; L = Lysostaphin]] | [[File:BBa_K2812004_secrety_lyso.png|900px|thumb|centre|Figure 1: Type I secretion of lysostaphin via HlyA, HlyB, HlyD and TolC. IM = inner membrane; OM = outer membrane; A = HlyA; L = Lysostaphin]] | ||
| − | ==Experimental Characterisation by TU Eindhoven (2018)== | + | ==Experimental Characterisation by TU-Eindhoven (2018)== |
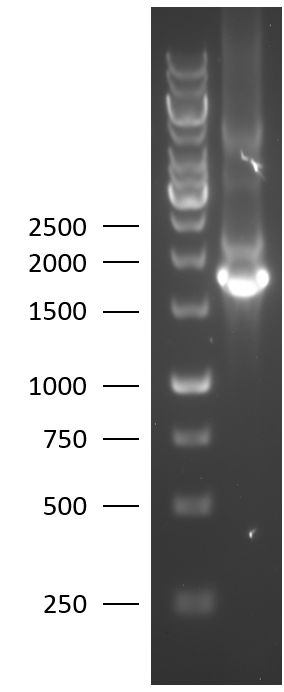
[[File:BBa_K2812004_Agarose_gel.png|125px|thumb|right|Figure 2: 1% agarose gel of the colony PCR of the Lysostaphin-HlyA biobrick.]] | [[File:BBa_K2812004_Agarose_gel.png|125px|thumb|right|Figure 2: 1% agarose gel of the colony PCR of the Lysostaphin-HlyA biobrick.]] | ||
===Cloning=== | ===Cloning=== | ||
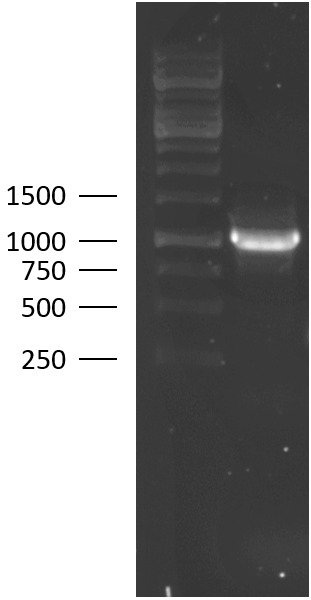
| − | TU Eindhoven 2018 has characterized the biobrick | + | TU-Eindhoven 2018 has characterized the biobrick <partinfo>BBa_K2812004</partinfo> at both the DNA and the protein level. First, the lysostaphin-thrombin linker-HlyA-His-tag construct was synthesized by IDT and subsequently double digested and assembled into the digested linearized pSB1C3 backbone via ligation. The ligated construct was successfully transformed into ''E. coli'' NovaBlue, followed by a colony PCR using the VF2 and VR primers to investigate if the correct length has been inserted in the vector. The mixture was ran on a 1% agarose gel as can be seen in figure 2. The observed length of the brightest band corresponds with the expected length of 1771 basepairs, confirming that the desired construct has been succesfully ligated in pSB1C3 and subsequently transformed in ''E. coli'' NovaBlue. Next, the colonies with the correct insert were cultured in LB before a plasmid purification by a miniprep. The isolated plasmid DNA was sent for Sanger sequencing and the sequence could be confirmed. |
===Protein Expression=== | ===Protein Expression=== | ||
[[File:BBa_K2812004+5_SDS_gel.png |125px|thumb|right|Figure 3: SDS-PAGE gel of the Lysostaphin-HlyA biobrick protein expression experiment.]] | [[File:BBa_K2812004+5_SDS_gel.png |125px|thumb|right|Figure 3: SDS-PAGE gel of the Lysostaphin-HlyA biobrick protein expression experiment.]] | ||
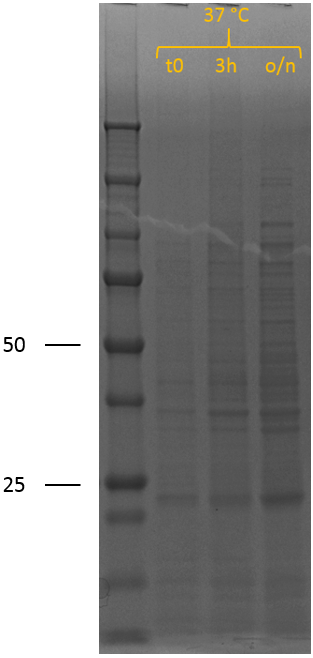
| − | After the successful characterisation of the biobrick at the DNA level, protein expression experiments were performed. To allow expression of the construct by the addition of IPTG, it was assembled behind the T7 promotor of iGEM Bielefeld 2011 <partinfo>BBa_K525998</partinfo> using restriction digestion by EcoRI and PstI followed by ligation. We have also included this construct as a new composite part in the registry, see <partinfo>BBa_K2812005</partinfo> for a more detailed characterisation at the DNA level. The biobrick was successfully transformed into BL21 (DE3), after which a culture was set up at 37 °C. A sample was taken | + | After the successful characterisation of the biobrick at the DNA level, protein expression experiments were performed. To allow expression of the construct by the addition of IPTG, it was assembled behind the T7 promotor of iGEM Bielefeld 2011 <partinfo>BBa_K525998</partinfo> using restriction digestion by EcoRI and PstI followed by ligation. We have also included this construct as a new composite part in the registry, see <partinfo>BBa_K2812005</partinfo> for a more detailed characterisation at the DNA level. The biobrick was successfully transformed into BL21 (DE3), after which a culture was set up at 37 °C. A sample was taken prior to induction (t0) to establish gene expression pattern of the uninduced bacteria. The cultures were induced at an OD600 of 0.5-0.8 by adding 0.5 mM IPTG to induce expression of the recombinant protein, also at 37 °C. Samples were taken 3 hours (3h) after induction and after overnight incubation (o/n). SDS samples were prepared and loaded onto a polyacrylamide gel to yield the SDS-PAGE results that can be seen in figure 3. |
| − | The expected length of the biobrick is 50 kDa. In the uninduced sample (t0), no bands indicative of overexpression of (any) protein can be observed. 3 hours after induction (3h) of protein expression, two bands indicating overexpression can be seen; one at 50 kDa and one at 25 kDa. After overnight induction, the band at 50 kDa has disappeared completely and the band around 25 kDa has increased in intensity. The SDS sample of 3h indicates successful induction of expression of the construct. The band at 25 kDa is not expected. The disappearance of the band at 50 kDa and increase of the 25 kDa band can be explained by cleavage of the thrombin linker due to accumulation of lysostaphin in the cytosol. The thrombin linker used in the design contains two times a GGGGS repeat. The substrate sequence of lysostaphin is GGGGG. However, it is known that the bacterial species S. simulans incorporates serine residues at the third and fifth position in the cell wall cross bridges (GGSGS), resulting in a 1000-fold decrease in susceptibility to lysostaphin as lysostaphin cannot hydrolyse glycylserine and serylglycine bonds.<sup>1</sup> Lysostaphin is thus still able to cleave these linkers, although at a lower rate. If the thrombin linker between lysostaphin and HlyA is cleaved by another lysostaphin enzyme, it yields two proteins of 27 kDa (lysostaphin) and 23 kDa (HlyA). The band around 25 kDa and the disappearance of the 50 kDa band can thus be explained by lysostaphin cleaving the thrombin linker. | + | The expected length of the biobrick is 50 kDa. In the uninduced sample (t0), no bands indicative of overexpression of (any) protein can be observed. 3 hours after induction (3h) of protein expression, two bands indicating overexpression can be seen; one at 50 kDa and one at 25 kDa. After overnight induction, the band at 50 kDa has disappeared completely and the band around 25 kDa has increased in intensity. The SDS sample of 3h indicates successful induction of expression of the construct. The band at 25 kDa is not expected. The disappearance of the band at 50 kDa and increase of the 25 kDa band can be explained by cleavage of the thrombin linker due to accumulation of lysostaphin in the cytosol. The thrombin linker used in the design contains two times a GGGGS repeat. The substrate sequence of lysostaphin is GGGGG. However, it is known that the bacterial species S. simulans incorporates serine residues at the third and fifth position in the cell wall cross bridges (GGSGS), resulting in a 1000-fold decrease in susceptibility to lysostaphin as lysostaphin cannot hydrolyse glycylserine and serylglycine bonds.<sup>1</sup> Lysostaphin is thus still able to cleave these linkers, although at a lower rate. If the thrombin linker between lysostaphin and HlyA is cleaved by another lysostaphin enzyme, it yields two proteins of 27 kDa (lysostaphin) and 23 kDa (HlyA). The band around 25 kDa and the disappearance of the 50 kDa band can thus be explained by lysostaphin cleaving the thrombin linker. From this experiment it can be concluded that our construct can be induced successfully. |
===Functionality Experiments=== | ===Functionality Experiments=== | ||
| Line 35: | Line 34: | ||
To prove that this cleavage does not occur rapidly intracellularly preventing secretion of the lysostaphin domain (as HlyA is lost), functional experiments were performed in collaboration with the PAMM foundation, which is the regional center of infectious diseases and pathology in South-East Brabant in the Netherlands. | To prove that this cleavage does not occur rapidly intracellularly preventing secretion of the lysostaphin domain (as HlyA is lost), functional experiments were performed in collaboration with the PAMM foundation, which is the regional center of infectious diseases and pathology in South-East Brabant in the Netherlands. | ||
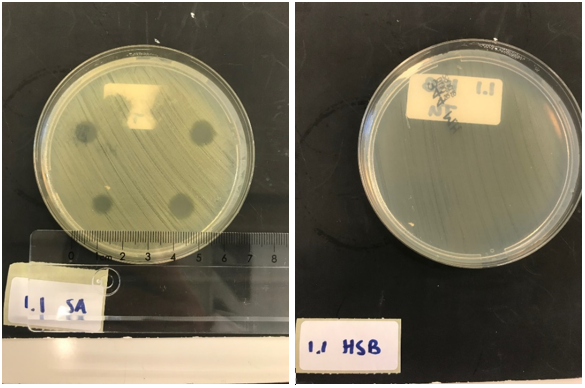
| − | To test the production and effectiveness of lysostaphin, BL21 (DE3) cells were transformed with the plasmid containing the T7 promotor from iGEM Bielefeld 2011 | + | To test the production and effectiveness of lysostaphin, BL21 (DE3) cells were transformed with the plasmid containing the T7 promotor from iGEM Bielefeld 2011 <partinfo>BBa_k525998</partinfo> assembled in front of this biobrick and subsequently cultured. Next, this culture was induced overnight. Only the production of lysostaphin is expected here, since the HlyB/D proteins required for type I secretion are missing. The cell lysate was taken from the culture and washed thoroughly, to remove any present antibiotic to exclude antibiotics as the source of bactericidal activity against ''S. aureus''. ''S. aureus'' and ''Streptococcus agalactiae'' were plated on separate Mueller-Hinton agar plates and 3 (left top and bottom) and 5 (right top and bottom) µL of ''E. coli'' cell lysate was applied. As a control, the same experiment was performed but now with a biofilm of ''Streptococcus agalactiae'' (HSB). This is another strain of gram positive bacteria that is not susceptible to lysostaphin but will be affected if e.g. traces of antibiotic are still present. |
After an overnight incubation with the cell lysate of our lysostaphin producing ''E. coli'', the results in figure 4 were obtained (3 µL top and bottom left; 5 µL top and bottom right): | After an overnight incubation with the cell lysate of our lysostaphin producing ''E. coli'', the results in figure 4 were obtained (3 µL top and bottom left; 5 µL top and bottom right): | ||
| Line 42: | Line 41: | ||
Clear halo formation on the ''S. aureus'' plate can be observed, indicating the production and activity of lysostaphin in the cell lysate. No halo formation was observed on the ''Streptococcus agalactiae'' plate, ’confirming that the results observed are caused by the lytic activity of lysostaphin produced by our ''E. coli'', and not by possible remains of antibiotics. | Clear halo formation on the ''S. aureus'' plate can be observed, indicating the production and activity of lysostaphin in the cell lysate. No halo formation was observed on the ''Streptococcus agalactiae'' plate, ’confirming that the results observed are caused by the lytic activity of lysostaphin produced by our ''E. coli'', and not by possible remains of antibiotics. | ||
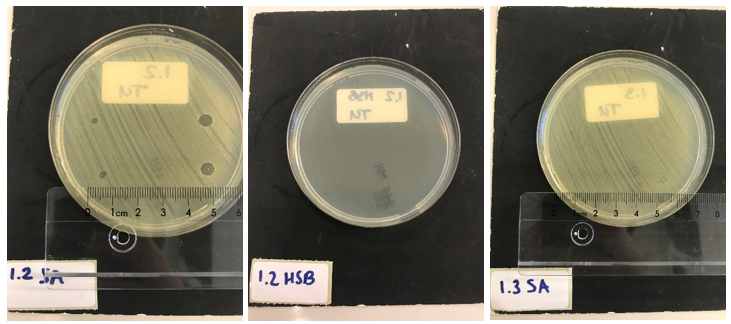
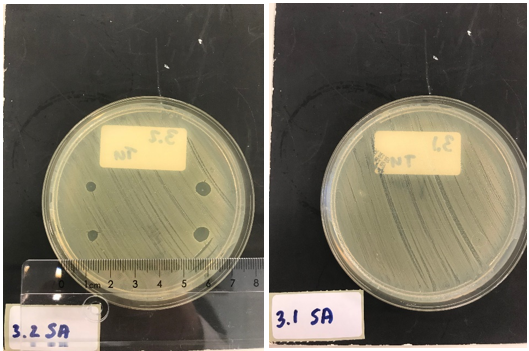
| − | Next, we tested the functionality of the secretion system. Since the pSB1C3 biobrick plasmid and the pSTV plasmid containting the HlyB/D secretion proteins had overlapping antibiotic resistances, the biobrick was assembled into a pBAD vector under control of an AraC promotor inducible by | + | Next, we tested the functionality of the secretion system. Since the pSB1C3 biobrick plasmid and the pSTV plasmid containting the HlyB/D secretion proteins had overlapping antibiotic resistances, the biobrick was assembled into a pBAD vector under control of an AraC promotor inducible by arabinose. ''E. coli'' BL21 (DE3) cells were transformed with both the HlyB/D plasmid and our newly assembled biobrick plasmid. Both plasmids were then simultaneously induced. Additionally, CaCl<sub>2</sub> was added, as this is required for Type I secretion. After four hours, the cultures were centrifuged and the cell pellets were lysed and washed. Similarly, the medium was filtered and washed extensively to remove any residual traces of antibiotics. A part of the medium was concentrated 10X and a part was concentrated 30X. As a control, BL21 (DE3) cells expressing both a modified construct without lysostaphin but only HlyA and the HlyB/D construct were tested. These bacteria can only secrete HlyA. After induction, samples from these bacteria were prepared according to the same protocol. ''S. aureus'' and ''Streptococcus agalactiae'' were plated on separate Mueller-Hinton agar plates to which 3 and 5 µL of ''E. coli'' cell lysate was applied. After an overnight incubation with the cell lysate of our lysostaphin producing ''E. coli'', the results in figure 5 were obtained (3 µL top and bottom left; 5 µL top and bottom right): |
| − | [[File:BBa_K2812004_pBAD_Lysate.png |500px|frame|centre|Figure 5: Left: ''S. Aureus'' plate with cell lysate of induced Lysostaphin-HlyA. Centre: ''Streptococcus agalactiae'' plate with cell lysate of induced Lysostaphin-HlyA. Right: ''S. Aureus'' plate with cell lysate of induced HlyA.]] | + | [[File:BBa_K2812004_pBAD_Lysate.png |500px|frame|centre|Figure 5: Left: ''S. Aureus'' plate with cell lysate of induced Lysostaphin-HlyA + induced HlyB/D. Centre: ''Streptococcus agalactiae'' plate with cell lysate of induced Lysostaphin-HlyA + induced HlyB/D. Right: ''S. Aureus'' plate with cell lysate of induced HlyA + induced HlyB/D.]] |
30x concentrated medium (Figure 6: 3 µL top and bottom left; 5 µL top and bottom right): | 30x concentrated medium (Figure 6: 3 µL top and bottom left; 5 µL top and bottom right): | ||
| − | [[File:BBa_K2812004_pBAD_Med_conc.png |500px|frame|centre|Figure 6: Left: ''S. aureus'' plate with 30X concentrated medium of induced Lysostaphin-HlyA. Centre: ''Streptococcus agalactiae'' plate with 30X concentrated medium of induced Lysostaphin-HlyA. | + | [[File:BBa_K2812004_pBAD_Med_conc.png |500px|frame|centre|Figure 6: Left: ''S. aureus'' plate with 30X concentrated medium of induced Lysostaphin-HlyA + induced HlyB/D. Centre: ''Streptococcus agalactiae'' plate with 30X concentrated medium of induced Lysostaphin-HlyA + induced HlyB/D. |
| − | Right: ''S. aureus'' plate with 30X concentrated medium of induced HlyA.]] | + | Right: ''S. aureus'' plate with 30X concentrated medium of induced HlyA + induced HlyB/D.]] |
10x concentrated medium (Figure 7: 3 µL top and bottom left; 5 µL top and bottom right): | 10x concentrated medium (Figure 7: 3 µL top and bottom left; 5 µL top and bottom right): | ||
| − | [[File:BBa_K2812004_pBAD_Med_dilute.png |500px|frame|centre|Figure 7: Left: ''S. aureus'' plate with 10X concentrated medium of induced Lysostaphin-HlyA. Right: ''S. aureus'' plate with 10X concentrated medium of induced HlyA.]] | + | [[File:BBa_K2812004_pBAD_Med_dilute.png |500px|frame|centre|Figure 7: Left: ''S. aureus'' plate with 10X concentrated medium of induced Lysostaphin-HlyA + induced HlyB/D. Right: ''S. aureus'' plate with 10X concentrated medium of induced HlyA + induced HlyB/D.]] |
Clear halo formation in 1.2 SA confirms that the lysostaphin in the cell lysate is active. Absence of halo in 1.2 HSB indicates that the lytic activity against ''S. aureus'' is not due to antibiotics remnants being present but has to be due to the Lysostaphin-HlyA construct. Absence of halo formation in 1.3 SA confirms that the lytic activity against ''S. aureus'' is due to lysostaphin, and not the HlyA domain. The same conclusion as the construct under control of the T7 promotor. | Clear halo formation in 1.2 SA confirms that the lysostaphin in the cell lysate is active. Absence of halo in 1.2 HSB indicates that the lytic activity against ''S. aureus'' is not due to antibiotics remnants being present but has to be due to the Lysostaphin-HlyA construct. Absence of halo formation in 1.3 SA confirms that the lytic activity against ''S. aureus'' is due to lysostaphin, and not the HlyA domain. The same conclusion as the construct under control of the T7 promotor. | ||
3.4 SA shows that the 30x concentrated medium of the induced Lysostaphin-HlyA has lytic acitivity against ''S. aureus'', while the HlyA construct 3.3 SA has no lytic acitivty. The medium of Lysostaphin-HlyA has no lytic acitivty against ''Streptococcus agalactiae'' (3.4 HSB). This confirms that lysostaphin is successfully secreted in the medium by our engineered ''E. coli'' bacteria and that the lytic activity observed against ''S. aureus'' is due to the lysostaphin and not HlyA or other components in the medium. Similar results are obtained with the 10X concentrated medium, showing a similar activity at 3X lower concentration. | 3.4 SA shows that the 30x concentrated medium of the induced Lysostaphin-HlyA has lytic acitivity against ''S. aureus'', while the HlyA construct 3.3 SA has no lytic acitivty. The medium of Lysostaphin-HlyA has no lytic acitivty against ''Streptococcus agalactiae'' (3.4 HSB). This confirms that lysostaphin is successfully secreted in the medium by our engineered ''E. coli'' bacteria and that the lytic activity observed against ''S. aureus'' is due to the lysostaphin and not HlyA or other components in the medium. Similar results are obtained with the 10X concentrated medium, showing a similar activity at 3X lower concentration. | ||
====Co-culture of lysostaphin-secreting ''E. coli'' and ''Staphylococcus aureus'' or ''Streptococcus agalactiae''==== | ====Co-culture of lysostaphin-secreting ''E. coli'' and ''Staphylococcus aureus'' or ''Streptococcus agalactiae''==== | ||
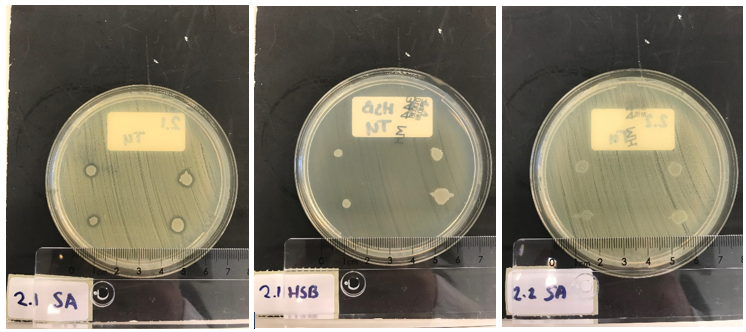
| − | Finally, as an ultimate experiment for confirming the secretion of the lysostaphin construct by ''E. coli'', a co-culture of ''S. aureus'' or ''Streptococcus agalactiae'' with ''E.coli'' BL21 (DE3) on Mueller-Hinton agar plates was performed. The plates were covered with IPTG, arabinose and CaCl<sub>2</sub> to induce our uninduced engineered ''E. coli'' bacteria to induce expression of lysostaphin-HlyA or HlyA and the HlyB/D secretion proteins. First ''S. aureus'' or ''Streptococcus agalactiae'' were plated on the inducer-agar. Subsequently, drops of uninduced BL21 (DE3) cultures were put on top of this film. The results can be seen in figure 8: | + | Finally, as an ultimate experiment for confirming the secretion of the lysostaphin construct by ''E. coli'', a co-culture of ''S. aureus'' or ''Streptococcus agalactiae'' with ''E. coli'' BL21 (DE3) on Mueller-Hinton agar plates was performed. The plates were covered with IPTG, arabinose and CaCl<sub>2</sub> to induce our uninduced engineered ''E. coli'' bacteria to induce expression of lysostaphin-HlyA or HlyA and the HlyB/D secretion proteins. First ''S. aureus'' or ''Streptococcus agalactiae'' were plated on the inducer-agar. Subsequently, drops of uninduced BL21 (DE3) cultures were put on top of this film. The results can be seen in figure 8: |
[[File:BBa_K2812004_pBAD_Cells.png |500px|frame|centre|Figure 8: Left: Co-culture of ''E. coli'' with induced lysostaphin-HlyA & HlyB/D expression and ''S. aureus''. Centre: Co-culture of ''E. coli'' with induced lysostaphin-HlyA & HlyB/D expression and ''Streptococcus agalactiae''. Right:: Co-culture of ''E. coli'' with induced HlyA & HlyB/D expression and ''S. Aureus''.]] | [[File:BBa_K2812004_pBAD_Cells.png |500px|frame|centre|Figure 8: Left: Co-culture of ''E. coli'' with induced lysostaphin-HlyA & HlyB/D expression and ''S. aureus''. Centre: Co-culture of ''E. coli'' with induced lysostaphin-HlyA & HlyB/D expression and ''Streptococcus agalactiae''. Right:: Co-culture of ''E. coli'' with induced HlyA & HlyB/D expression and ''S. Aureus''.]] | ||
| − | In 2.1 SA, clear halo formation around the BL21 (DE3) colonies can be observed, indicating secretion of lysostaphin having a lytic effect on ''S. aureus''. 2.1 HSB clearly shows that on another Gram positive bacterial strain, lysostaphin does not show any antimicrobial activity. As a final control, BL21 (DE3) only expressing HlyA and HlyB/D (without lysostaphin), was co-cultured with ''S. aureus'' (see 2.2 SA). Around these ''E.coli'' colonies, no halo formation can be observed again illustrating that the lysostaphin component of the fusion proteins contains the lytic effect selectively against ''S. aureus'', and HlyA is not toxic. | + | In 2.1 SA, clear halo formation around the BL21 (DE3) colonies can be observed, indicating secretion of lysostaphin having a lytic effect on ''S. aureus''. 2.1 HSB clearly shows that on another Gram positive bacterial strain, lysostaphin does not show any antimicrobial activity. As a final control, BL21 (DE3) only expressing HlyA and HlyB/D (without lysostaphin), was co-cultured with ''S. aureus'' (see 2.2 SA). Around these ''E. coli'' colonies, no halo formation can be observed again illustrating that the lysostaphin component of the fusion proteins contains the lytic effect selectively against ''S. aureus'', and HlyA is not toxic. |
| − | [[File:BBa_K2812004_Stockholm2016_ColonyPCR.png|125px|thumb|right|Figure | + | ====Co-culture of lysostaphin-secreting ''E. coli'' and Methicillin-resistant ''Staphylococcus aureus''==== |
| − | [[File:BBa_K2812004_Stockholm2016.png|125px|thumb|right|Figure | + | Additionally, to illustrate the power of lysostaphin, the exact same co-culture experiment was conducted on a biofilm of methicillin-resistant ''S. aureus'' (MRSA). MRSA was plated on the plates already containing the inducers (IPTG and arabinose) and CaCl2. The ''E. coli'' BL21 (DE3) bacteria containing the lysostaphin-HlyA or HlyA and the HlyB/D contructs were loaded on top of the MRSA covered plates. The results can be seen in figure 9: |
| + | [[File:BBa_K2812004_coculture_MRSA.png|750px|thumb|centre|Figure 9: Left: Co-culture of ''E. coli'' with induced lysostaphin-HlyA & HlyB/D expression and Methicillin-resistant ''S. aureus''. Right:: Co-culture of ''E. coli'' with induced HlyA & HlyB/D expression and Methicillin-resistant ''S. Aureus''.]] | ||
| + | The same observations were made as in the other co-culture experiment. In 1.4, clear halo formation around the BL21 (DE3) colonies can be observed, indicating secretion of lysostaphin having a lytic effect on S. aureus. 1.6 clearly shows that BL21 (DE3) only expressing HlyA and HlyB/D (without lysostaphin) does not show any antimicrobial activity. Around these E.coli colonies, no halo formation can be observed again illustrating that the lysostaphin component of the fusion proteins contains the lytic effect selectively against S. aureus, and HlyA is not toxic. | ||
| + | |||
| + | [[File:BBa_K2812004_Stockholm2016_ColonyPCR.png|125px|thumb|right|Figure 10: 1% agarose gel of the colony PCR of the T7-truncated lysostaphin construct.]] | ||
| + | [[File:BBa_K2812004_Stockholm2016.png|125px|thumb|right|Figure 11: SDS-PAGE of the truncated lysostaphin construct under control of T7 promotor. First row: BioRad All Blue Protein ladder, t0 is uninduced sample, 3h and o/n is time after induction.]] | ||
====Functional improvement of <partinfo>BBa_K748002</partinfo>==== | ====Functional improvement of <partinfo>BBa_K748002</partinfo>==== | ||
| − | To demonstrate the functional improvement over the original truncated lysostaphin <partinfo>BBa_K748002</partinfo> created by HIT-Harbin 2012, we intended to assemble our biobrick | + | To demonstrate the functional improvement over the original truncated lysostaphin <partinfo>BBa_K748002</partinfo> created by HIT-Harbin 2012, we intended to assemble our biobrick <partinfo>BBa_K2812004</partinfo> with the T7 promotor <partinfo>BBa_k525998</partinfo>, allowing protein expression by induction with IPTG. Since the part <partinfo>BBa_K2144001</partinfo> from iGEM Stockholm 2016 is this exact assembly, we requested their part from the iGEM registry and we transformed it successfully in ''E. coli'' BL21 (DE3). Colony PCR was used to pick colonies carrying the correct insert length. The results can be seen in figure 10. After subsequent culturing and miniprep to isolate the plasmid DNA, we sent the sample for sanger sequencing and could confirm the sequence. |
| − | Transformed ''E. coli'' BL21 (DE3) bacteria were cultured overnight at 37 °C, followed by protein induction at an OD600 of 0.5-0.8 by adding 0.5 mM IPTG, also at 37 °C. Before induction, a cell pellet sample was taken as control (uninduced SDS sample, t0). Cell pellet samples were collected after 3 hours of induction (induced SDS sample, 3h) and after overnight induction (induced SDS sample, o/n). All samples were prepared for SDS-PAGE. The SDS gel can be seen in figure | + | Transformed ''E. coli'' BL21 (DE3) bacteria were cultured overnight at 37 °C, followed by protein induction at an OD600 of 0.5-0.8 by adding 0.5 mM IPTG, also at 37 °C. Before induction, a cell pellet sample was taken as control (uninduced SDS sample, t0). Cell pellet samples were collected after 3 hours of induction (induced SDS sample, 3h) and after overnight induction (induced SDS sample, o/n). All samples were prepared for SDS-PAGE. The SDS gel can be seen in figure 11. No clear overexpression of a band around 27 kDa, the expected length of truncated lysostaphin, can be seen after induction of the construct. There is no difference, except for overall intensity due to bacterial growth, between the to and 3h sample. Similarly, the o/n sample shows no difference with the samples at t0 or 3h, except for overall intensity due to bacterial growth. As no convincing expression of the sequence confirmed construct can be observed, our codon optimisation improved the expression of the lysostaphin domain considerably. However, in our eyes the most valuable functional improvement of this part over <partinfo>BBa_K748002</partinfo> is the ability of lysostaphin to be secreted, which is obviously not possible for the <partinfo>BBa_K748002</partinfo> part lacking the HlyA domain. |
===Overall conclusion=== | ===Overall conclusion=== | ||
| − | We have demonstrated that the designed | + | We have demonstrated that the designed lysostaphin-HlyA fusion protein gets successfully secreted by ''E. coli'' via Type I secretion if HlyB/D is co-expressed in the bacteria. The lysostaphin-HlyA biobrick has a lytic effect against ''S. aureus'' and the construct is not toxic for other, non-susceptible bacteria like ''Streptococcus agalactiae''. Importantly, the construct is not active against ''E. coli'' and secretion enables continuous release. Additionally, the use of cell lysis to release lysostaphin is no longer required, functionally improving the truncated lysostaphin construct from iGEM Harbin 2012 which required cell lysis for release in their system. Besides release, isolation of lysostaphin with this newly designed construct is also easier as purification can be performed directly from the medium, avoiding the requirement of cell lysis and decreasing purification problems. |
Unexpectedly, self-cleavage of the thrombin linker is observed if lysostaphin is allowed to concentrate due to cleavage at the GGGGS sites of the linker. To minimize self-cleavage, partial inhibition of lysostaphin can be achieved by adding excess Zinc to the culture medium.<sup>1</sup> Alternatively, the design of the linker could be changed to avoid repeats of glycine residues. | Unexpectedly, self-cleavage of the thrombin linker is observed if lysostaphin is allowed to concentrate due to cleavage at the GGGGS sites of the linker. To minimize self-cleavage, partial inhibition of lysostaphin can be achieved by adding excess Zinc to the culture medium.<sup>1</sup> Alternatively, the design of the linker could be changed to avoid repeats of glycine residues. | ||
<!-- --> | <!-- --> | ||
| Line 74: | Line 78: | ||
1) Tossavainen, H., Raulinaitis, V., Kauppinen, L., Pentikäinen, U., Maaheimo, H., & Permi, P. (2018). Structural and Functional Insights Into Lysostaphin–Substrate Interaction. ''Front Mol Biosci''. | 1) Tossavainen, H., Raulinaitis, V., Kauppinen, L., Pentikäinen, U., Maaheimo, H., & Permi, P. (2018). Structural and Functional Insights Into Lysostaphin–Substrate Interaction. ''Front Mol Biosci''. | ||
| − | 2) Bastos, M. d., Coutinho, B. G., & Coelho, M. L. (2010). Lysostaphin: A Staphylococcal Bacteriolysin with Potential Clinical Applications. Pharmaceuticals (Basel) , 1139–1161. | + | 2) Bastos, M. d., Coutinho, B. G., & Coelho, M. L. (2010). Lysostaphin: A Staphylococcal Bacteriolysin with Potential Clinical Applications. ''Pharmaceuticals (Basel)'', 1139–1161. |
| − | 3) Thomas, S., Holland, I. B., & Schmitt, L. (2014). The Type 1 secretion pathway — The hemolysin system and beyond. Molecular Cell Research, 1629-1641. | + | 3) Thomas, S., Holland, I. B., & Schmitt, L. (2014). The Type 1 secretion pathway — The hemolysin system and beyond. ''Molecular Cell Research'', 1629-1641. |
| − | 4) Gray, L., Baker, K., Kenny, B., Mackman, N., Haigh, R., & Holland, I. (1989). A novel C-terminal signal sequence targets ''Escherichia coli'' haemolysin directly to the medium. J Cell Sci Suppl., 45-57. | + | 4) Gray, L., Baker, K., Kenny, B., Mackman, N., Haigh, R., & Holland, I. (1989). A novel C-terminal signal sequence targets ''Escherichia coli'' haemolysin directly to the medium. ''J Cell Sci Suppl.'', 45-57. |
| − | 5) Wandersman, C., & Delepelaire, P. (1990 ). TolC, an ''Escherichia coli'' outer membrane protein required for hemolysin secretion. Proc Natl Acad Sci U S A. , 4776-4780. | + | 5) Wandersman, C., & Delepelaire, P. (1990 ). TolC, an ''Escherichia coli'' outer membrane protein required for hemolysin secretion. ''Proc Natl Acad Sci U S A.'', 4776-4780. |
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
Latest revision as of 03:55, 18 October 2018
Coding sequence for trunctated Lysostaphin fused to His-tagged HlyA
The biobrick contains the coding domain for truncated lysostaphin, based on the coding sequence derived from BBa_K748002. This was fused to the C-terminal sequence of Hemolysin A (HlyA) via a thrombin linker to allow secretion of the lysostaphin. TU-Eindhoven 2018 used this part to secrete lysostaphin from Escherichia coli to kill Staphylococcus aureus biofilms for the treatment of wound infections. For more information about our project, please visit our [http://2018.igem.org/Team:TU-Eindhoven wiki].
Usage & Biology
Lysostaphin
Lysostaphin is an antimicrobial agent produced by Staphylococcus simulans. It targets the cell wall peptidoglycan found in certain Staphylococci by cleaving its cross-linking pentaglycine bridges. Among others, it is effective for degrading Staphylococcus aureus biofilms.1 The encoding part of the lysostaphin has been derived from BBa_K748002, which was made by iGEM Harbin 2012 and was also used by iGEM Stockholm 2016. iGEM Eindhoven 2018 codon optimized this lysostaphin construct. Lysostaphin belongs to the major class of antimicrobial proteins and peptides known as bacteriocins. Bacteriocins are proteins or peptides produced by bacteria, displaying a bactericidal activity against other subpopulations of bacteria.2 The cell wall degradation capability of lysostaphin derives from its endopeptidase activity on pentaglycine cross-bridges in the peptidoglycan layer. Specific cleavage between the third and fourth glycine residue leads to the destruction of the peptidoglycan layer and subsequent lysis of the bacteria.
HlyA
The C-terminal sequence of Hemolysin A (residues 807-1024 of the E. coli HlyA gene) functions as a non-cleavable signal peptide for protein translocation via the Type I secretion pathway of Gram-negative bacteria. Here, HlyA is fused to the lysostaphin for its secretion from E. coli BL21 (DE3) cells. In Type I secretion, single step transport of the target protein occurs from the cytoplasm to the extracellular environment. The HlyA is fused to the C-terminus of the target protein.3 It is a 23-kDa signal sequence that targets proteins for secretion via Type I secretion pathway. HlyA is secreted into the medium in a TolC and HlyB/D-dependent manner, in the presence of CaCl2.4 It is recognized by the membrane translocation complex composed of HlyB and HlyD, which together with the TolC protein, will form a pore through the membrane.5 This will lead to the secretion of HlyA-containing fusion proteins. Figure 1 illustrates the steps involved in the type I secretion of lysostaphin as an HlyA fusion protein.
Thrombin linker & His-tag
The lysostaphin and HlyA are linked via a thrombin linker. It is a short peptide sequence which can be cleaved by the enzyme thrombin, resulting in the removal of the HlyA domain. This avoids interference with the functionality of lysostaphin. C-terminal to the HlyA domain, a His-tag (6x repeated amino acid histidine) is attached. The His-tag can be used for detection of the fusion protein by e.g. Western Blot, but also for protein purification purposes as it facilitates binding to a nickel affinity column.
Fusion protein
The combination of all components described above, lead to a construct that can be (continuously) secreted by E. coli if the HlyB/D coding sequences have been co-transformed. Running the medium containing the fusion protein over a nickel affinity column and subsequent thrombin linker cleavage allows for the easy isolation of purified truncated lysostaphin.
Experimental Characterisation by TU-Eindhoven (2018)
Cloning
TU-Eindhoven 2018 has characterized the biobrick BBa_K2812004 at both the DNA and the protein level. First, the lysostaphin-thrombin linker-HlyA-His-tag construct was synthesized by IDT and subsequently double digested and assembled into the digested linearized pSB1C3 backbone via ligation. The ligated construct was successfully transformed into E. coli NovaBlue, followed by a colony PCR using the VF2 and VR primers to investigate if the correct length has been inserted in the vector. The mixture was ran on a 1% agarose gel as can be seen in figure 2. The observed length of the brightest band corresponds with the expected length of 1771 basepairs, confirming that the desired construct has been succesfully ligated in pSB1C3 and subsequently transformed in E. coli NovaBlue. Next, the colonies with the correct insert were cultured in LB before a plasmid purification by a miniprep. The isolated plasmid DNA was sent for Sanger sequencing and the sequence could be confirmed.
Protein Expression
After the successful characterisation of the biobrick at the DNA level, protein expression experiments were performed. To allow expression of the construct by the addition of IPTG, it was assembled behind the T7 promotor of iGEM Bielefeld 2011 BBa_K525998 using restriction digestion by EcoRI and PstI followed by ligation. We have also included this construct as a new composite part in the registry, see BBa_K2812005 for a more detailed characterisation at the DNA level. The biobrick was successfully transformed into BL21 (DE3), after which a culture was set up at 37 °C. A sample was taken prior to induction (t0) to establish gene expression pattern of the uninduced bacteria. The cultures were induced at an OD600 of 0.5-0.8 by adding 0.5 mM IPTG to induce expression of the recombinant protein, also at 37 °C. Samples were taken 3 hours (3h) after induction and after overnight incubation (o/n). SDS samples were prepared and loaded onto a polyacrylamide gel to yield the SDS-PAGE results that can be seen in figure 3.
The expected length of the biobrick is 50 kDa. In the uninduced sample (t0), no bands indicative of overexpression of (any) protein can be observed. 3 hours after induction (3h) of protein expression, two bands indicating overexpression can be seen; one at 50 kDa and one at 25 kDa. After overnight induction, the band at 50 kDa has disappeared completely and the band around 25 kDa has increased in intensity. The SDS sample of 3h indicates successful induction of expression of the construct. The band at 25 kDa is not expected. The disappearance of the band at 50 kDa and increase of the 25 kDa band can be explained by cleavage of the thrombin linker due to accumulation of lysostaphin in the cytosol. The thrombin linker used in the design contains two times a GGGGS repeat. The substrate sequence of lysostaphin is GGGGG. However, it is known that the bacterial species S. simulans incorporates serine residues at the third and fifth position in the cell wall cross bridges (GGSGS), resulting in a 1000-fold decrease in susceptibility to lysostaphin as lysostaphin cannot hydrolyse glycylserine and serylglycine bonds.1 Lysostaphin is thus still able to cleave these linkers, although at a lower rate. If the thrombin linker between lysostaphin and HlyA is cleaved by another lysostaphin enzyme, it yields two proteins of 27 kDa (lysostaphin) and 23 kDa (HlyA). The band around 25 kDa and the disappearance of the 50 kDa band can thus be explained by lysostaphin cleaving the thrombin linker. From this experiment it can be concluded that our construct can be induced successfully.
Functionality Experiments
Cell lysates & Medium
To prove that this cleavage does not occur rapidly intracellularly preventing secretion of the lysostaphin domain (as HlyA is lost), functional experiments were performed in collaboration with the PAMM foundation, which is the regional center of infectious diseases and pathology in South-East Brabant in the Netherlands.
To test the production and effectiveness of lysostaphin, BL21 (DE3) cells were transformed with the plasmid containing the T7 promotor from iGEM Bielefeld 2011 BBa_K525998 assembled in front of this biobrick and subsequently cultured. Next, this culture was induced overnight. Only the production of lysostaphin is expected here, since the HlyB/D proteins required for type I secretion are missing. The cell lysate was taken from the culture and washed thoroughly, to remove any present antibiotic to exclude antibiotics as the source of bactericidal activity against S. aureus. S. aureus and Streptococcus agalactiae were plated on separate Mueller-Hinton agar plates and 3 (left top and bottom) and 5 (right top and bottom) µL of E. coli cell lysate was applied. As a control, the same experiment was performed but now with a biofilm of Streptococcus agalactiae (HSB). This is another strain of gram positive bacteria that is not susceptible to lysostaphin but will be affected if e.g. traces of antibiotic are still present.
After an overnight incubation with the cell lysate of our lysostaphin producing E. coli, the results in figure 4 were obtained (3 µL top and bottom left; 5 µL top and bottom right):
Clear halo formation on the S. aureus plate can be observed, indicating the production and activity of lysostaphin in the cell lysate. No halo formation was observed on the Streptococcus agalactiae plate, ’confirming that the results observed are caused by the lytic activity of lysostaphin produced by our E. coli, and not by possible remains of antibiotics.
Next, we tested the functionality of the secretion system. Since the pSB1C3 biobrick plasmid and the pSTV plasmid containting the HlyB/D secretion proteins had overlapping antibiotic resistances, the biobrick was assembled into a pBAD vector under control of an AraC promotor inducible by arabinose. E. coli BL21 (DE3) cells were transformed with both the HlyB/D plasmid and our newly assembled biobrick plasmid. Both plasmids were then simultaneously induced. Additionally, CaCl2 was added, as this is required for Type I secretion. After four hours, the cultures were centrifuged and the cell pellets were lysed and washed. Similarly, the medium was filtered and washed extensively to remove any residual traces of antibiotics. A part of the medium was concentrated 10X and a part was concentrated 30X. As a control, BL21 (DE3) cells expressing both a modified construct without lysostaphin but only HlyA and the HlyB/D construct were tested. These bacteria can only secrete HlyA. After induction, samples from these bacteria were prepared according to the same protocol. S. aureus and Streptococcus agalactiae were plated on separate Mueller-Hinton agar plates to which 3 and 5 µL of E. coli cell lysate was applied. After an overnight incubation with the cell lysate of our lysostaphin producing E. coli, the results in figure 5 were obtained (3 µL top and bottom left; 5 µL top and bottom right):
30x concentrated medium (Figure 6: 3 µL top and bottom left; 5 µL top and bottom right):
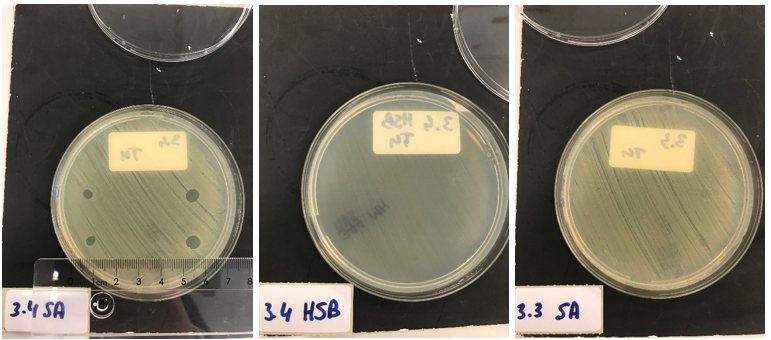
10x concentrated medium (Figure 7: 3 µL top and bottom left; 5 µL top and bottom right):
Clear halo formation in 1.2 SA confirms that the lysostaphin in the cell lysate is active. Absence of halo in 1.2 HSB indicates that the lytic activity against S. aureus is not due to antibiotics remnants being present but has to be due to the Lysostaphin-HlyA construct. Absence of halo formation in 1.3 SA confirms that the lytic activity against S. aureus is due to lysostaphin, and not the HlyA domain. The same conclusion as the construct under control of the T7 promotor. 3.4 SA shows that the 30x concentrated medium of the induced Lysostaphin-HlyA has lytic acitivity against S. aureus, while the HlyA construct 3.3 SA has no lytic acitivty. The medium of Lysostaphin-HlyA has no lytic acitivty against Streptococcus agalactiae (3.4 HSB). This confirms that lysostaphin is successfully secreted in the medium by our engineered E. coli bacteria and that the lytic activity observed against S. aureus is due to the lysostaphin and not HlyA or other components in the medium. Similar results are obtained with the 10X concentrated medium, showing a similar activity at 3X lower concentration.
Co-culture of lysostaphin-secreting E. coli and Staphylococcus aureus or Streptococcus agalactiae
Finally, as an ultimate experiment for confirming the secretion of the lysostaphin construct by E. coli, a co-culture of S. aureus or Streptococcus agalactiae with E. coli BL21 (DE3) on Mueller-Hinton agar plates was performed. The plates were covered with IPTG, arabinose and CaCl2 to induce our uninduced engineered E. coli bacteria to induce expression of lysostaphin-HlyA or HlyA and the HlyB/D secretion proteins. First S. aureus or Streptococcus agalactiae were plated on the inducer-agar. Subsequently, drops of uninduced BL21 (DE3) cultures were put on top of this film. The results can be seen in figure 8:
In 2.1 SA, clear halo formation around the BL21 (DE3) colonies can be observed, indicating secretion of lysostaphin having a lytic effect on S. aureus. 2.1 HSB clearly shows that on another Gram positive bacterial strain, lysostaphin does not show any antimicrobial activity. As a final control, BL21 (DE3) only expressing HlyA and HlyB/D (without lysostaphin), was co-cultured with S. aureus (see 2.2 SA). Around these E. coli colonies, no halo formation can be observed again illustrating that the lysostaphin component of the fusion proteins contains the lytic effect selectively against S. aureus, and HlyA is not toxic.
Co-culture of lysostaphin-secreting E. coli and Methicillin-resistant Staphylococcus aureus
Additionally, to illustrate the power of lysostaphin, the exact same co-culture experiment was conducted on a biofilm of methicillin-resistant S. aureus (MRSA). MRSA was plated on the plates already containing the inducers (IPTG and arabinose) and CaCl2. The E. coli BL21 (DE3) bacteria containing the lysostaphin-HlyA or HlyA and the HlyB/D contructs were loaded on top of the MRSA covered plates. The results can be seen in figure 9:
The same observations were made as in the other co-culture experiment. In 1.4, clear halo formation around the BL21 (DE3) colonies can be observed, indicating secretion of lysostaphin having a lytic effect on S. aureus. 1.6 clearly shows that BL21 (DE3) only expressing HlyA and HlyB/D (without lysostaphin) does not show any antimicrobial activity. Around these E.coli colonies, no halo formation can be observed again illustrating that the lysostaphin component of the fusion proteins contains the lytic effect selectively against S. aureus, and HlyA is not toxic.
Functional improvement of BBa_K748002
To demonstrate the functional improvement over the original truncated lysostaphin BBa_K748002 created by HIT-Harbin 2012, we intended to assemble our biobrick BBa_K2812004 with the T7 promotor BBa_K525998, allowing protein expression by induction with IPTG. Since the part BBa_K2144001 from iGEM Stockholm 2016 is this exact assembly, we requested their part from the iGEM registry and we transformed it successfully in E. coli BL21 (DE3). Colony PCR was used to pick colonies carrying the correct insert length. The results can be seen in figure 10. After subsequent culturing and miniprep to isolate the plasmid DNA, we sent the sample for sanger sequencing and could confirm the sequence.
Transformed E. coli BL21 (DE3) bacteria were cultured overnight at 37 °C, followed by protein induction at an OD600 of 0.5-0.8 by adding 0.5 mM IPTG, also at 37 °C. Before induction, a cell pellet sample was taken as control (uninduced SDS sample, t0). Cell pellet samples were collected after 3 hours of induction (induced SDS sample, 3h) and after overnight induction (induced SDS sample, o/n). All samples were prepared for SDS-PAGE. The SDS gel can be seen in figure 11. No clear overexpression of a band around 27 kDa, the expected length of truncated lysostaphin, can be seen after induction of the construct. There is no difference, except for overall intensity due to bacterial growth, between the to and 3h sample. Similarly, the o/n sample shows no difference with the samples at t0 or 3h, except for overall intensity due to bacterial growth. As no convincing expression of the sequence confirmed construct can be observed, our codon optimisation improved the expression of the lysostaphin domain considerably. However, in our eyes the most valuable functional improvement of this part over BBa_K748002 is the ability of lysostaphin to be secreted, which is obviously not possible for the BBa_K748002 part lacking the HlyA domain.
Overall conclusion
We have demonstrated that the designed lysostaphin-HlyA fusion protein gets successfully secreted by E. coli via Type I secretion if HlyB/D is co-expressed in the bacteria. The lysostaphin-HlyA biobrick has a lytic effect against S. aureus and the construct is not toxic for other, non-susceptible bacteria like Streptococcus agalactiae. Importantly, the construct is not active against E. coli and secretion enables continuous release. Additionally, the use of cell lysis to release lysostaphin is no longer required, functionally improving the truncated lysostaphin construct from iGEM Harbin 2012 which required cell lysis for release in their system. Besides release, isolation of lysostaphin with this newly designed construct is also easier as purification can be performed directly from the medium, avoiding the requirement of cell lysis and decreasing purification problems. Unexpectedly, self-cleavage of the thrombin linker is observed if lysostaphin is allowed to concentrate due to cleavage at the GGGGS sites of the linker. To minimize self-cleavage, partial inhibition of lysostaphin can be achieved by adding excess Zinc to the culture medium.1 Alternatively, the design of the linker could be changed to avoid repeats of glycine residues.
Sources
1) Tossavainen, H., Raulinaitis, V., Kauppinen, L., Pentikäinen, U., Maaheimo, H., & Permi, P. (2018). Structural and Functional Insights Into Lysostaphin–Substrate Interaction. Front Mol Biosci.
2) Bastos, M. d., Coutinho, B. G., & Coelho, M. L. (2010). Lysostaphin: A Staphylococcal Bacteriolysin with Potential Clinical Applications. Pharmaceuticals (Basel), 1139–1161.
3) Thomas, S., Holland, I. B., & Schmitt, L. (2014). The Type 1 secretion pathway — The hemolysin system and beyond. Molecular Cell Research, 1629-1641.
4) Gray, L., Baker, K., Kenny, B., Mackman, N., Haigh, R., & Holland, I. (1989). A novel C-terminal signal sequence targets Escherichia coli haemolysin directly to the medium. J Cell Sci Suppl., 45-57.
5) Wandersman, C., & Delepelaire, P. (1990 ). TolC, an Escherichia coli outer membrane protein required for hemolysin secretion. Proc Natl Acad Sci U S A., 4776-4780.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1351
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 126
- 1000COMPATIBLE WITH RFC[1000]