Difference between revisions of "Part:BBa K2559005"
| (5 intermediate revisions by one other user not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K2559005 short</partinfo> | <partinfo>BBa_K2559005 short</partinfo> | ||
| − | The BBa_K2559005 is a | + | <p>The [https://parts.igem.org/Part:BBa_K2559005# BBa_K2559005] is a amended eGFP coding part improved from [https://parts.igem.org/Part:BBa_l714891# BBa_l714891]. |
===Usage and Biology=== | ===Usage and Biology=== | ||
| − | The part BBa_K2559006 has | + | The part BBa_K2559006 has a sequence improvement on the basic part submitted by iGEM07_Peking ([https://parts.igem.org/Part:BBa_I714891# BBa_I714891]) which encodes the SDY_eGFP. However, we found out a 16 bp nucleotides redundancy in the eGFP starting coding region in BBa_I714891, after checking the sequence of BBa_I714891 from NCBI. Therefore, we decided to delete the redundant 16 bp nucleotides in BBa_I714891 to amend the length of eGFP coding sequence. The amended eGFP coding biobrick is the BBa_K2559005. |
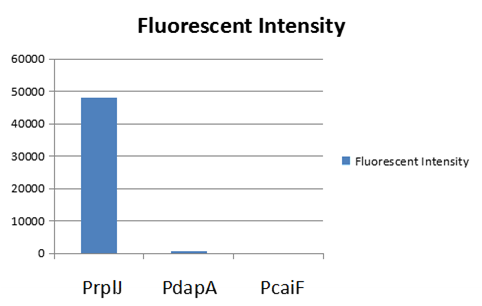
| − | + | To test the function of BBa_K2559005, we designed a new E.coli expression vector containing our new part termed as [https://parts.igem.org/Part:BBa_K2559003# BBa_K2559003]under a strong E.coli endogenous promoter (PrplJ). Therefore, the amended eGFP in BBa_K2559005 was driven by PrplJ promoter, and expressed in DH10B. In addition, we also applied the BBa_K2559005 in the promoter intensity analysis of our other two new parts, the [https://parts.igem.org/Part:BBa_K2559004# BBa_K2559004] and [https://parts.igem.org/Part:BBa_K2559011# BBa_K2559011] which are relatively weaker E.coli endogenous promoters (PdapA and PcaiF) (Figure 1). | |
| + | |||
| + | [[File:Scau-china-2018-11.png|800px|thumb|center]] | ||
| + | |||
| + | |||
| + | [[File:Scau-china-2018-12.png|800px|thumb|center|Figure 1: Fluorescent intensity of amended eGFP driven by by PrplJ, PdapA, PcaiF promoter. | ||
| + | ]] | ||
| + | |||
| + | |||
| + | |||
| + | We summarized that our improvedpart, the amended eGFP coding biobrick BBa_K2559005 worked well in DH10B. We also hoped that our improvement on the BBa_I714891 can help their future applications by other groups in the future. However, it is difficult for us to perform additional experiments with BBa_K2559005 and BBa_I714891 due to the unavailable BBa_I714891. | ||
| + | |||
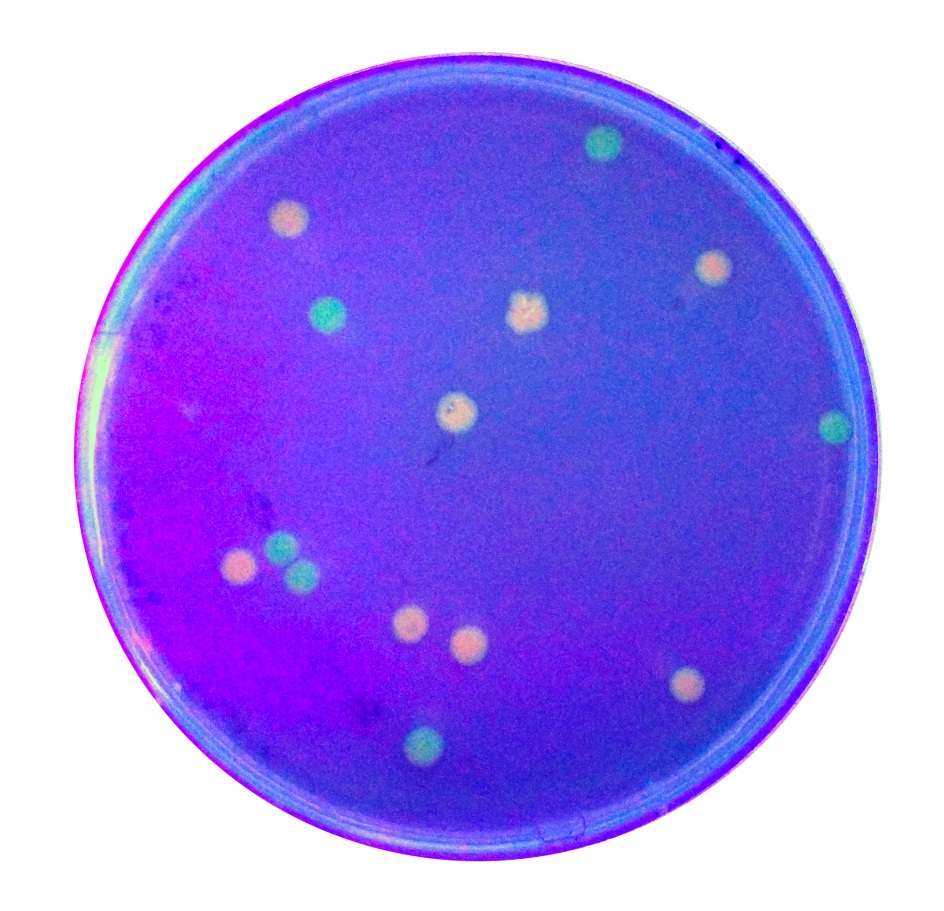
| + | To expand the application of BBa_K2559005, we searched the[https://parts.igem.org/Part:BBa_J04450# BBa_J04450]stored in registry and do another improvement in the BBa_J04450. The BBa_J04450 is a strong RFP expression vector in E.coli. As the main page of BBa_J04450 mentioned, the E.coli colonies with BBa_J04450 were in red color under normal light after about 18 hour culture on LB plate (Figure 2). We used the BBa_K2559005 to replace the RFP region in BBa_J04450, the modified part is [https://parts.igem.org/Part:BBa_K2559009# BBa_K2559009]. | ||
| + | We transferred the BBa_K2559009 to DH5α by heat-shock, and found that the fluorescence signal can be observed under the UV (Figure 2). | ||
| + | |||
| + | [[File:T-SCAU-China-Red_and_green_colonies.jpg|200px|thumb|center|Figure 2 : Figure 2 Colonies with BBa_J04450 (red colony) and BBa_K2559009 (green colony) were visualized under UV lightbox | ||
| + | ]] | ||
| + | |||
| + | So, we confirm that our improved part BBa_K2559005 can work in different E.coli expression system. We are also looking forward to more application of the BBa_K2559009! | ||
| + | |||
| + | == JiangnanU_China 2019 == | ||
| + | |||
| + | In 2018, the team iGEM18_SCAU-China chose a strong E. coli endogenous promoter (PrplJ,BBa_K2559003) to express their amended eGFP (BBa_K2559005). This year our team JiangnanU_China also chose this promoter PrplJ (BBa_K2559003) to express their amended eGFP (BBa_K2559005). The team iGEM18_SCAU-China chose one timepoint to test the promoter strength. However, we measured the value of (fluorescence intensity)/OD600 at 0 h, 4 h, 6 h, 8 h and 10 h. Through the change of value what we measured, we can found the promoter PrplJ worked well. In the other hand, we observed the eGFP fluorescent protein by fluorescence microscope and we found that the E. coli with the amended eGFP fluorescent protein emitted the noticeable green fluorescence. | ||
| + | |||
| + | [[Image:PrplJ-eGFP-1.png|700px|thumb|center|Figure1. The curve of Fluorescent intensity/OD600 - Time(h)]] | ||
| + | |||
| + | [[Image:PrplJ-eGFP-2.png|650px|thumb|center|Figure2. Fluorescent intensity of eGFP driven by by PrplJ promoter]] | ||
| − | |||
Latest revision as of 08:57, 19 October 2019
Amended eGFP
The BBa_K2559005 is a amended eGFP coding part improved from BBa_l714891.
Usage and Biology
The part BBa_K2559006 has a sequence improvement on the basic part submitted by iGEM07_Peking (BBa_I714891) which encodes the SDY_eGFP. However, we found out a 16 bp nucleotides redundancy in the eGFP starting coding region in BBa_I714891, after checking the sequence of BBa_I714891 from NCBI. Therefore, we decided to delete the redundant 16 bp nucleotides in BBa_I714891 to amend the length of eGFP coding sequence. The amended eGFP coding biobrick is the BBa_K2559005. To test the function of BBa_K2559005, we designed a new E.coli expression vector containing our new part termed as BBa_K2559003under a strong E.coli endogenous promoter (PrplJ). Therefore, the amended eGFP in BBa_K2559005 was driven by PrplJ promoter, and expressed in DH10B. In addition, we also applied the BBa_K2559005 in the promoter intensity analysis of our other two new parts, the BBa_K2559004 and BBa_K2559011 which are relatively weaker E.coli endogenous promoters (PdapA and PcaiF) (Figure 1).
We summarized that our improvedpart, the amended eGFP coding biobrick BBa_K2559005 worked well in DH10B. We also hoped that our improvement on the BBa_I714891 can help their future applications by other groups in the future. However, it is difficult for us to perform additional experiments with BBa_K2559005 and BBa_I714891 due to the unavailable BBa_I714891.
To expand the application of BBa_K2559005, we searched theBBa_J04450stored in registry and do another improvement in the BBa_J04450. The BBa_J04450 is a strong RFP expression vector in E.coli. As the main page of BBa_J04450 mentioned, the E.coli colonies with BBa_J04450 were in red color under normal light after about 18 hour culture on LB plate (Figure 2). We used the BBa_K2559005 to replace the RFP region in BBa_J04450, the modified part is BBa_K2559009. We transferred the BBa_K2559009 to DH5α by heat-shock, and found that the fluorescence signal can be observed under the UV (Figure 2).
So, we confirm that our improved part BBa_K2559005 can work in different E.coli expression system. We are also looking forward to more application of the BBa_K2559009!
JiangnanU_China 2019
In 2018, the team iGEM18_SCAU-China chose a strong E. coli endogenous promoter (PrplJ,BBa_K2559003) to express their amended eGFP (BBa_K2559005). This year our team JiangnanU_China also chose this promoter PrplJ (BBa_K2559003) to express their amended eGFP (BBa_K2559005). The team iGEM18_SCAU-China chose one timepoint to test the promoter strength. However, we measured the value of (fluorescence intensity)/OD600 at 0 h, 4 h, 6 h, 8 h and 10 h. Through the change of value what we measured, we can found the promoter PrplJ worked well. In the other hand, we observed the eGFP fluorescent protein by fluorescence microscope and we found that the E. coli with the amended eGFP fluorescent protein emitted the noticeable green fluorescence.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]