Difference between revisions of "Part:BBa K2637010"
| (9 intermediate revisions by the same user not shown) | |||
| Line 4: | Line 4: | ||
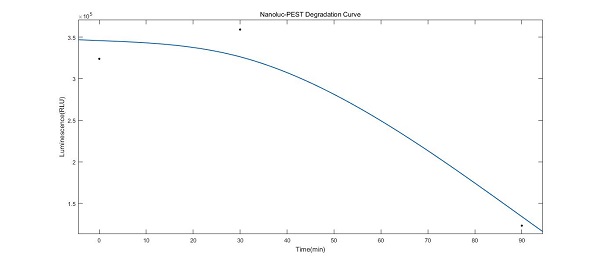
| − | This | + | This biobrick contais two parts. The first one is Nanoluc luciferase, which is from iGEM2015_Tuebingen [https://parts.igem.org/Part:BBa_K1680009 BBa_K1680009]. The NanoLuc is smaller than other luciferases while yielding equal or stronger luminescence readings. We have optimized it in yeast. The second part is a tag named PEST. PEST sequence can be found in variety of eukaryotic cells and the sequence we used was from the literature Engineered Luciferase Reporter from a Deep Sea Shrimp Utilizing a Novel Imidazopyrazinone Substrate. The covalent linkage of ubiquitin to lysine residues of substrate proteins is a common means used by eucaryotic cells to signal their degradation by the 26S proteasome, a multiprotease complex located in the cytoplasm and the nucleus. Decades ago it had been demonstrated that the PEST regions, enriched with Pro, Glu, Ser, and Thr, were identified to indeed control the ubiquitination of regulatory short-lived proteins. |
| + | |||
| + | [[Image:ganggangnano.jpeg|center|frame|300px|<b>Figure 1. </b> Nanoluc-PEST Degradation Curve]] | ||
| + | |||
| + | Inspired by our time-course measurement for the KaiABC oscillatory system, we realized the importance of the sensitivity of reporter genes when used to characterize the variation in our engineering strains. | ||
| + | With the knowledge that the instability of proteins is associated with the existence of the so-called PEST regions, which control the ubiquitination of regulatory short-lived proteins, we decided to optimize our NanoLuc with the potentially degrading sequence PSET to shorten its intracellular lifetime. | ||
| + | |||
| + | As shown in the data image, the ''NanoLuc'' was expressed and degraded successfully as we expected which embodied that the mark PEST also worked. | ||
Our basic parts range from BBa_K2637001 to BBa_K2637018 and BBa_K2637053 to BBa_K2637059. | Our basic parts range from BBa_K2637001 to BBa_K2637018 and BBa_K2637053 to BBa_K2637059. | ||
| Line 32: | Line 39: | ||
==Overview of Cyanobacterias' circadian rhythm== | ==Overview of Cyanobacterias' circadian rhythm== | ||
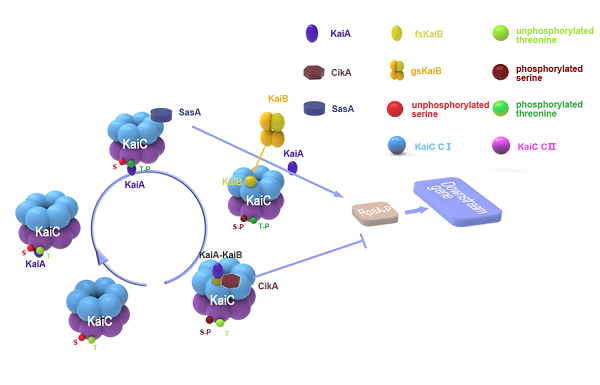
| − | [[Image:ganggangm3.png|center|frame|300px|<b>Figure | + | [[Image:ganggangm3.png|center|frame|300px|<b>Figure 2. </b>Mechanism figure of our project]] |
Organisms are adapted to the relentless cycles of day and night, because they evolved timekeeping systems called circadian clocks, which regulate biological activities with ~24-hour rhythms. The clock of cyanobacteria is driven by a three-protein oscillator composed of only three protein KaiA, KaiB and KaiC, which together generate a circadian rhythm of KaiC phosphorylation at residues serine 431 and threonine 432 in the CII dimain. KaiA promotes KaiC (auto)phosphorylation during the subjective day, whereas KaiB provides inhibition of KaiA and promotes KaiC (auto)dephosphorylation during the subjective night. The 24-h KaiC phosphorylation pattern can be reconstituted in vitro by merely combining the three Kai proteins and ATP, suggesting that It is post-transcriptional oscillations and is only related to proteins. Like KaiA, KaiB is also involved in regulating two antagonistic clock-output proteins--SasA and CikA, which reciprocally control the master regulator of transcription RpaA. | Organisms are adapted to the relentless cycles of day and night, because they evolved timekeeping systems called circadian clocks, which regulate biological activities with ~24-hour rhythms. The clock of cyanobacteria is driven by a three-protein oscillator composed of only three protein KaiA, KaiB and KaiC, which together generate a circadian rhythm of KaiC phosphorylation at residues serine 431 and threonine 432 in the CII dimain. KaiA promotes KaiC (auto)phosphorylation during the subjective day, whereas KaiB provides inhibition of KaiA and promotes KaiC (auto)dephosphorylation during the subjective night. The 24-h KaiC phosphorylation pattern can be reconstituted in vitro by merely combining the three Kai proteins and ATP, suggesting that It is post-transcriptional oscillations and is only related to proteins. Like KaiA, KaiB is also involved in regulating two antagonistic clock-output proteins--SasA and CikA, which reciprocally control the master regulator of transcription RpaA. | ||
| Line 40: | Line 47: | ||
| − | [[Image:ganggangusage.jpeg|center|frame|300px|<b>Figure | + | [[Image:ganggangusage.jpeg|center|frame|300px|<b>Figure 3. </b> Usage of Life Tik Tok]] |
Life Tik Tok is a circadian rhythm that is established in yeast by iGEM Tianjin, 2018. It enables to regular the biological activities in yeast under the control of proteins in KaiABC oscillator. Learn more about Life Tik Tok and click here.[http://2018.igem.org/Team:Tianjin] | Life Tik Tok is a circadian rhythm that is established in yeast by iGEM Tianjin, 2018. It enables to regular the biological activities in yeast under the control of proteins in KaiABC oscillator. Learn more about Life Tik Tok and click here.[http://2018.igem.org/Team:Tianjin] | ||
| Line 73: | Line 80: | ||
=Characterization of Life Tik Tok= | =Characterization of Life Tik Tok= | ||
| − | ==Characterization of | + | ==Characterization of Life Tik Tok== |
| − | + | ||
| − | + | ||
| − | + | When we start to measure the datas of experiment group, we set up some control groups at the same time. To prove that our oscillator works surely because of the yeast two-hybrid system, we constructed some bacteria in which there are only one fusion protein of yeast two-hybrid system and a reporter gene or a combination. And we choose 3 or 4 reporter genes as parallel experiment groups. | |
| + | The following figures are the results we measured with live Nanoluc. | ||
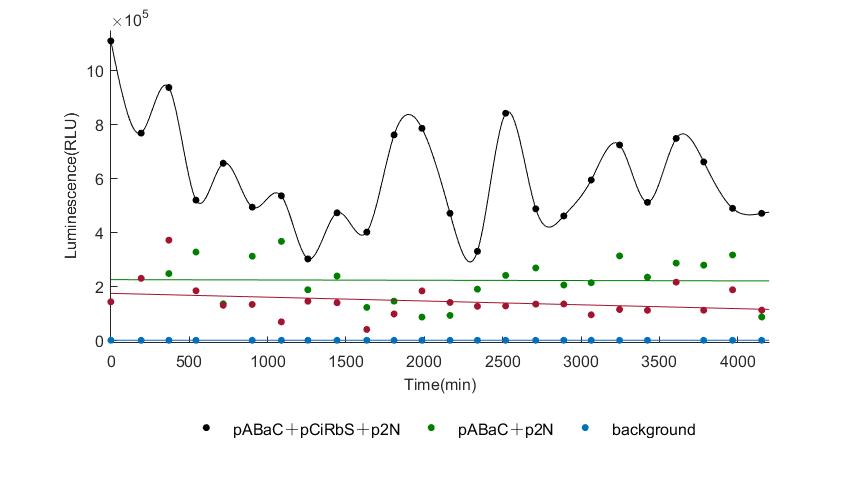
| + | [[Image:gangganglive1.jpeg|center|frame|300px|<b>Figure 4. </b> The result of oscillator measured by the 1st live Nanoluc reporter together with the control groups]] | ||
| − | + | [[Image:gangganglive2.jpeg|center|frame|300px|<b>Figure 5. </b> The result of oscillator measured by the 1st live Nanoluc reporter subtracting the 1st control groups]] | |
| − | + | [[Image:gangganglive3.jpeg|center|frame|300px|<b>Figure 6. </b> The result of oscillator measured by the 1st live Nanoluc reporter subtracting the 2nd control groups]] | |
| − | [[Image: | + | [[Image:gangganglive4.jpeg|center|frame|300px|<b>Figure 7. </b> The result of oscillator measured by the 2nd live Nanoluc reporter together with the control groups]] |
| − | [[Image: | + | [[Image:gangganglive5.jpeg|center|frame|300px|<b>Figure 8. </b> The result of oscillator measured by the 2nd live Nanoluc reporter subtracting the 1st control groups]] |
| − | [[Image: | + | [[Image:gangganglive6.jpeg|center|frame|300px|<b>Figure 9. </b> The result of oscillator measured by the 2nd live Nanoluc reporter subtracting the 2nd control groups]] |
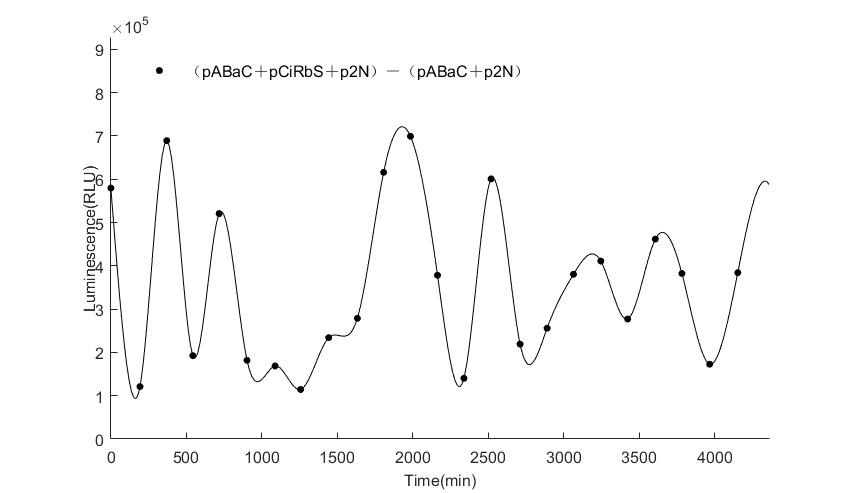
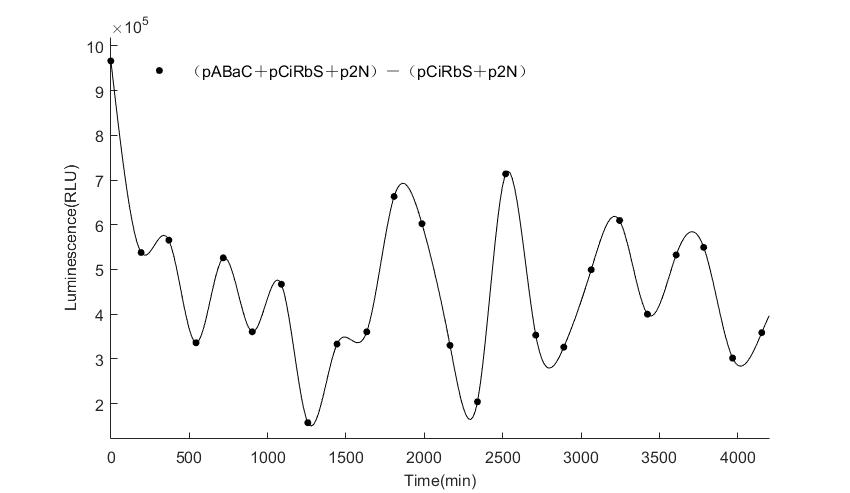
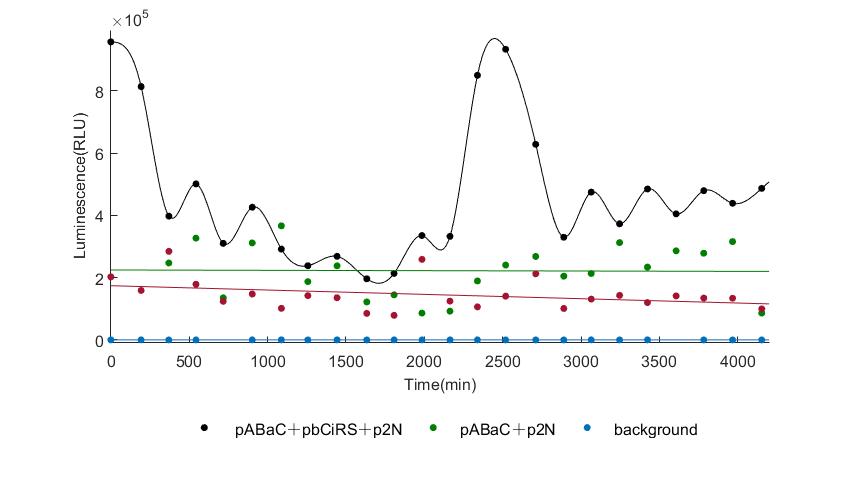
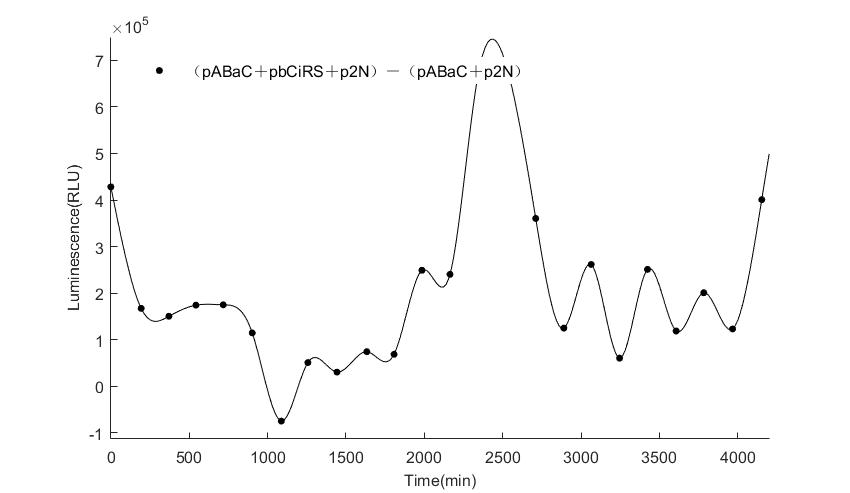
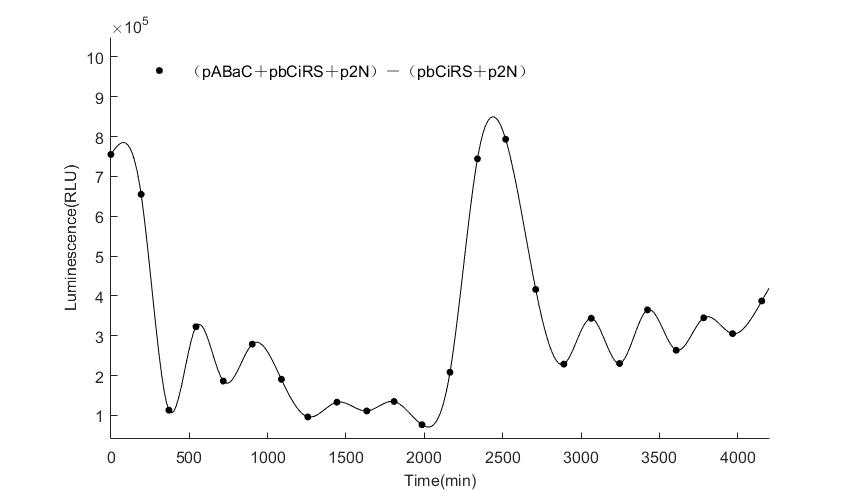
| − | In the | + | In the figures4-6, pABaC is the plasmid with ''AD-KaiC'' fused gene and make up a yeast two-hybrid system with the plasmid |
| − | + | pCiRbS, which is with ''BD-CikA'' fused gene. p1F is the plasmid with Nanoluc reporter whose promoter is ''Gal2''. In Figure.4, the experimental group have black datas and the first control group and the second group have green and red datas, respectively. We substracted the control groups to gat a more objective result. | |
As we predicted and looked at the results in other literatures, this oscillatory system is unstable.However, even though the fluctuations are not obvious on the second day, better waveforms could be observed on the first and third days.In addition, on the third day we get very high peak values, and in the wave trough experimental report gene expression is closer to the control group and the blank group, this indicates that in subjective day, yeast two-hybrid system works and results the peak value, in subjective night, it doesn't work and causes the similar results to the control groups and the background. | As we predicted and looked at the results in other literatures, this oscillatory system is unstable.However, even though the fluctuations are not obvious on the second day, better waveforms could be observed on the first and third days.In addition, on the third day we get very high peak values, and in the wave trough experimental report gene expression is closer to the control group and the blank group, this indicates that in subjective day, yeast two-hybrid system works and results the peak value, in subjective night, it doesn't work and causes the similar results to the control groups and the background. | ||
Therefore, the overall experimental results show that we successfully reconstructed the oscillator in yeast. | Therefore, the overall experimental results show that we successfully reconstructed the oscillator in yeast. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
You can see more details of our results [http://2018.igem.org/Team:Tianjin/Demonstrate here] | You can see more details of our results [http://2018.igem.org/Team:Tianjin/Demonstrate here] | ||
| − | |||
=References= | =References= | ||
Latest revision as of 13:49, 15 October 2018
Nanoluc+PEST
This biobrick contais two parts. The first one is Nanoluc luciferase, which is from iGEM2015_Tuebingen BBa_K1680009. The NanoLuc is smaller than other luciferases while yielding equal or stronger luminescence readings. We have optimized it in yeast. The second part is a tag named PEST. PEST sequence can be found in variety of eukaryotic cells and the sequence we used was from the literature Engineered Luciferase Reporter from a Deep Sea Shrimp Utilizing a Novel Imidazopyrazinone Substrate. The covalent linkage of ubiquitin to lysine residues of substrate proteins is a common means used by eucaryotic cells to signal their degradation by the 26S proteasome, a multiprotease complex located in the cytoplasm and the nucleus. Decades ago it had been demonstrated that the PEST regions, enriched with Pro, Glu, Ser, and Thr, were identified to indeed control the ubiquitination of regulatory short-lived proteins.
Inspired by our time-course measurement for the KaiABC oscillatory system, we realized the importance of the sensitivity of reporter genes when used to characterize the variation in our engineering strains. With the knowledge that the instability of proteins is associated with the existence of the so-called PEST regions, which control the ubiquitination of regulatory short-lived proteins, we decided to optimize our NanoLuc with the potentially degrading sequence PSET to shorten its intracellular lifetime.
As shown in the data image, the NanoLuc was expressed and degraded successfully as we expected which embodied that the mark PEST also worked.
Our basic parts range from BBa_K2637001 to BBa_K2637018 and BBa_K2637053 to BBa_K2637059. Our composite parts range from BBa_K2637021 to BBa_K2637044 and BBa_K2637047 to BBa_K2637052. Our part collection include all our basic parts except for BBa_K2637018 and all composite parts.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Biology and Usage
Overview of Cyanobacterias' circadian rhythm
Organisms are adapted to the relentless cycles of day and night, because they evolved timekeeping systems called circadian clocks, which regulate biological activities with ~24-hour rhythms. The clock of cyanobacteria is driven by a three-protein oscillator composed of only three protein KaiA, KaiB and KaiC, which together generate a circadian rhythm of KaiC phosphorylation at residues serine 431 and threonine 432 in the CII dimain. KaiA promotes KaiC (auto)phosphorylation during the subjective day, whereas KaiB provides inhibition of KaiA and promotes KaiC (auto)dephosphorylation during the subjective night. The 24-h KaiC phosphorylation pattern can be reconstituted in vitro by merely combining the three Kai proteins and ATP, suggesting that It is post-transcriptional oscillations and is only related to proteins. Like KaiA, KaiB is also involved in regulating two antagonistic clock-output proteins--SasA and CikA, which reciprocally control the master regulator of transcription RpaA.
Usage of Life Tik Tok (Cyanobacterias' circadian rhythm in yeast)
Life Tik Tok is a circadian rhythm that is established in yeast by iGEM Tianjin, 2018. It enables to regular the biological activities in yeast under the control of proteins in KaiABC oscillator. Learn more about Life Tik Tok and click here.[http://2018.igem.org/Team:Tianjin]
Intereaction between the proteins
Stepwise binding of two KaiA dimers triggers KaiC autophosphorylation at Thr432 and Ser431 . These phosphorylation events enable cooperative binding of fold-switched KaiB monomers to the KaiC-CI domain, forming the KaiCB complex. KaiCB provides a scaffold for the successive sequestration of KaiA in ternary KaiCBA assemblies, concurring with a rearrangement of the KaiA PsR domains. KaiA sequestration promotes KaiC autodephosphorylation, resulting in the regeneration of free KaiC through release of KaiBA subcomplexes. Temporal information from the oscillator is transmitted to downstream genes via the histidine protein kinase SasA (Synechococcus adaptive sensor A), whose autophosphorylation is stimulated by interaction with KaiC. Phosphorylated SasA in turn transfers a phosphoryl group to RpaA (regulator of phycobilisome association A) , a transcription factor that directly regulates the expression of approximately 100 genes. Moreover, RpaA indirectly regulates the expression of nearly all genes in the genome. Disruption of sasA also results in severely damped gene expression rhythms. Surprisingly, the phosphorylation state of RpaA, and subsequently its activity, have been shown to be dependent on CikA, which was primarily thought to be involved in entrainment.
Reconstruction of Life Tik Tok in yeast
When we reconstruct the KaiABC oscillator in yeast, our goals are:
·Characterize the combination between KaiABC changing over time via yeast two-hybrid system
·Explore the role of other proteins in stabilizing the oscillation without involving TTFL
·Discuss the effect of KaiC concentration on oscillation and select the proper promoters
·Research how heterologous circadian clock influences chromosome structure in yeast
We aim to integrate the KaiABC system into Saccharomyces cerevisiae BY4741, and have it control circadian rhythm in yeast. To do this, we have designed three plasmids which can be transformed into yeast to produce a oscillation system and characterized the protein interactions by yeast two-hybrid system. You can learn something about yeast two-hybrid system by clicking here. The first plasmid expresses KaiA, KaiB and KaiC which is fused to a Gal4 activation domain. The second plasmid is our reporter plasmid, which has fluorescent protein promoted by mutant Gal1p and luciferase promoted by Gal2p. The third plasmid contains CikA, SasA and RpaA. We respectively link a Gal4 binding domain with CikA or SasA to find the suitable binding protein that can characterize the oscillation. To explore more protein interactions, KaiB and KaiC were also used as proteins in the yeast two-hybrid system to construct a new oscillation system.These systems would conclusively show that the KaiABC system can work normally in yeast, which serves as a proof of concept for placing eukaryotic gene expression under the control of an exogenous circadian clock. In addition, to make sure that Gal1p and Gal2p can only be activited after the combination of proteins, we deleted two genes in wild-type BY4741. They are Gal4 and Gal80, which can activate or repress Gal1p and Gal2p, respectively. The principle is explained in yeast two-hybrid. And then, we deleted gene BarI to solve the problem of desynchronization between different generations of yeast.
Moreover, we detects the strength of several common promoters in yeast to find the proper one that provides suitable concentration. Thanks to our modeling, we can calculate the range of concentration of KaiC to support the oscillation, which contributes a lot to our selection of promoters.
Characterization of Life Tik Tok
Characterization of Life Tik Tok
When we start to measure the datas of experiment group, we set up some control groups at the same time. To prove that our oscillator works surely because of the yeast two-hybrid system, we constructed some bacteria in which there are only one fusion protein of yeast two-hybrid system and a reporter gene or a combination. And we choose 3 or 4 reporter genes as parallel experiment groups. The following figures are the results we measured with live Nanoluc.
In the figures4-6, pABaC is the plasmid with AD-KaiC fused gene and make up a yeast two-hybrid system with the plasmid pCiRbS, which is with BD-CikA fused gene. p1F is the plasmid with Nanoluc reporter whose promoter is Gal2. In Figure.4, the experimental group have black datas and the first control group and the second group have green and red datas, respectively. We substracted the control groups to gat a more objective result. As we predicted and looked at the results in other literatures, this oscillatory system is unstable.However, even though the fluctuations are not obvious on the second day, better waveforms could be observed on the first and third days.In addition, on the third day we get very high peak values, and in the wave trough experimental report gene expression is closer to the control group and the blank group, this indicates that in subjective day, yeast two-hybrid system works and results the peak value, in subjective night, it doesn't work and causes the similar results to the control groups and the background. Therefore, the overall experimental results show that we successfully reconstructed the oscillator in yeast.
You can see more details of our results [http://2018.igem.org/Team:Tianjin/Demonstrate here]
References
[1]Roger Tseng, Nicolette F. Goularte, Archana Chavan, Jansen Luu, Susan E. Cohen, Yong-Gang Chang, Joel Heisler, Sheng Li, Alicia K. Michael, Sarvind Tripathi, Susan S. Golden, Andy LiWang, Carrie L. Partch, Structural basis of the day-night transition in a bacterial circadian clock. Science, 1174-1180 (2017).
[2]Joost Snijder, Jan M. Schuller, Anika Wiegard, Philip Lössl, Nicolas Schmelling, Ilka M. Axmann, Jürgen M. Plitzko, Friedrich Förster, Albert J. R. Heck, Structures of the cyanobacterial circadian oscillator frozen in a fully assembled state. Science, 1181-1184 (2017).
[3]Joseph S.Markson, Joseph R.Piechura, Anna M.Puszynska, Erin K.O’Shea, Circadian Control of Global Gene Expression by the Cyanobacterial Master Regulator RpaA. Cell, 1396-1408 (2013).
[4]Jun O. Liu,et al.Everything you need to know about the yeast two-hybrid system.[J]. Nature Structural Biology, 1998, 535-536
[5]Hideo Iwasaki,* Stanly B. Hideo, Iwasaki. Sensory Histidine Kinase, SasA, Necessary to Sustain Robust Circadian Oscillation in Cyanobacteria.[J]. cell, 2000, 101(2): 223-233