Difference between revisions of "Part:BBa K2559011"
(→Usage and Biology) |
|||
| (3 intermediate revisions by 2 users not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K2559011 short</partinfo> | <partinfo>BBa_K2559011 short</partinfo> | ||
| − | + | The BBa_K2559004 , PcaiF promoter is a weak endogenous promoter in Escherichia coli K 12. | |
| − | + | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| + | We obtained this promoter from a database named PromEC ( http://margalit.huji.ac.il/promec/index.html) .PromEC is an updated compilation of E. coli mRNA promoter sequences. It includes detailed information of genesites which have been experimentally identified mRNA transcriptional starting position in the E. coli chromosome, as well as the actual sequences of the promoters. | ||
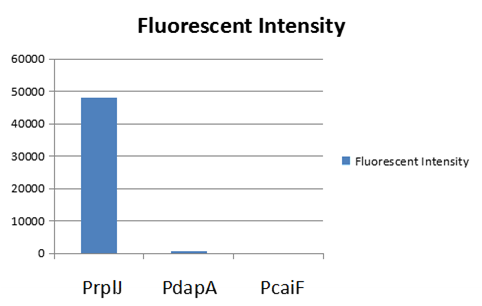
| + | We have built up a model to measure the expression intensity of these promoters. Eventually, we selected three promoters including: PrplJ, Pdapa and PcaiF based on the results of modeling PrplJ, Pdapa and PcaiF.We further made chimeric fluorescent fusion proteins between the promoter and GFP respectively and tested the fluorescent intensity driven by different promoters. The combination of PromEC and our modeling can be used as a efficient tool to figure out the information about interested promoters. The GFP expression in E.coli (DH5 α) and was driven by PrplJ, Pdapa and PcaiF promoter. | ||
| − | + | [[File:Scau-china-2018-11.png|800px|thumb|center|Figure 1 The result of the fluorescent intensity measurement ]] | |
| + | [[File:Scau-china-2018-12.png|800px|thumb|center|Figure 2 Fluorescent intensity of eGFP driven by by rplj/dapa/caif promoter.]] | ||
| + | The indensity of PcaiF is very weak and we hardly can observe the GFP fluorescent signal. | ||
| + | The part is facilitate the in-depth research for other teams! | ||
| + | |||
| + | |||
| + | <!-- --> | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
| − | <partinfo> | + | <partinfo>BBa_K2559004 SequenceAndFeatures</partinfo> |
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
===Functional Parameters=== | ===Functional Parameters=== | ||
| − | <partinfo> | + | <partinfo>BBa_K2559004 parameters</partinfo> |
<!-- --> | <!-- --> | ||
Latest revision as of 18:13, 15 October 2018
PcaiF promoter
The BBa_K2559004 , PcaiF promoter is a weak endogenous promoter in Escherichia coli K 12.
Usage and Biology
We obtained this promoter from a database named PromEC ( http://margalit.huji.ac.il/promec/index.html) .PromEC is an updated compilation of E. coli mRNA promoter sequences. It includes detailed information of genesites which have been experimentally identified mRNA transcriptional starting position in the E. coli chromosome, as well as the actual sequences of the promoters. We have built up a model to measure the expression intensity of these promoters. Eventually, we selected three promoters including: PrplJ, Pdapa and PcaiF based on the results of modeling PrplJ, Pdapa and PcaiF.We further made chimeric fluorescent fusion proteins between the promoter and GFP respectively and tested the fluorescent intensity driven by different promoters. The combination of PromEC and our modeling can be used as a efficient tool to figure out the information about interested promoters. The GFP expression in E.coli (DH5 α) and was driven by PrplJ, Pdapa and PcaiF promoter.
The indensity of PcaiF is very weak and we hardly can observe the GFP fluorescent signal.
The part is facilitate the in-depth research for other teams!
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]