Difference between revisions of "Part:BBa K2703008"
Saniya kari (Talk | contribs) |
Chlamy Dima (Talk | contribs) |
||
| (3 intermediate revisions by one other user not shown) | |||
| Line 6: | Line 6: | ||
=='''Introduction'''== | =='''Introduction'''== | ||
| − | For the silver medal criteria “Validated Part” we submit a basic part: the sequence of resistance marker to paromomycine that is an aminoglycoside antibiotic. The aphVIII gene from Streptomyces rimosus encode for an aminoglycoside 3’-phosphotransferase gene (aph). From the aphVIII gene sequence of the BioBrick BBa_K1884013 (iGEM16_SZU-China) | + | For the silver medal criteria “Validated Part” we submit a basic part: the sequence of resistance marker to paromomycine that is an aminoglycoside antibiotic. The aphVIII gene from Streptomyces rimosus encode for an aminoglycoside 3’-phosphotransferase gene (aph). From the aphVIII gene sequence of the BioBrick BBa_K1884013 (iGEM16_SZU-China) BpiI estriction site were removed ( G551C) by PCR point mutation. |
We cloned aphVIII by PCR amplification in B4-B5 position of the MoClo standard2 to use it in our retrotransposon design. Acceptor plasmid is the MoClo Universal pL0 acceptor plasmid pAGM91212. We sequenced again the sequence to be sure there is no unwanted mutations. | We cloned aphVIII by PCR amplification in B4-B5 position of the MoClo standard2 to use it in our retrotransposon design. Acceptor plasmid is the MoClo Universal pL0 acceptor plasmid pAGM91212. We sequenced again the sequence to be sure there is no unwanted mutations. | ||
| Line 78: | Line 78: | ||
</table> | </table> | ||
<figcaption>Figure 2: The 4 different devices made to test the functionality of the promoter PPSAD. The construction were made with the MoClo kit made for C.reinhardtii by P.Crozet et al 2018 1. </figcaption> | <figcaption>Figure 2: The 4 different devices made to test the functionality of the promoter PPSAD. The construction were made with the MoClo kit made for C.reinhardtii by P.Crozet et al 2018 1. </figcaption> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/parts/d/dc/T--Sorbonne_U_Paris--Bba_K2703008_data.png"> | ||
| + | <figcaption>Figure 3: Graphic listing the 4 devices (pCM-1) transformed in C.reinhardtii D66 by electroporation with 100 ng of DNA. The selection was made on TAP agar media with paromomycin (15 g/mL) by plate. Data are mean SD (N=3) and there were no colonies with a negative control .</figcaption> | ||
The aphVIII gene sequence work as intented in once a transcription unit is assembled by GoldenGate cloning following the Chlamydomonas MoClo kit. | The aphVIII gene sequence work as intented in once a transcription unit is assembled by GoldenGate cloning following the Chlamydomonas MoClo kit. | ||
| + | </html> | ||
| + | |||
| + | =='''Improvement'''== | ||
| + | <html> | ||
| + | |||
| + | The Humboldt Team 2019 proposed an improvement of this part for the gold criterium #2. The improved part, BBa_K2984006, can be seen <a href="https://parts.igem.org/Part:BBa_K2984006">here</a>. The part was improved by doing a codon optimization for use in <i>Chlamydomonas reinhardtii</i>. | ||
| + | |||
| + | <h3>Improvement: Methods </h3> | ||
| + | |||
| + | To see if the expression of the aminoglycoside 3’-phosphotransferase was increased, we performed several electroporations to transform C. reinhardtii with the improved paromomycin resistance. We used the C.reinhardtii strain UVM 4 since it is a strain designed to express transgene constructs (Neupert et al. 2009). We transformed the two paromomycin constructs with standard and improved codon usage starting with 0,5 µg DNA per electroporation sample and ascended with 0,5 µg steps up to 2 µg. For each construct and DNA mass we did three electroporations. The electroporation electrical resistance was measured for each sample. After resuspension and one day recovery in TAP medium, all samples were plated on TAP-agar plates containing a paromomycin concentration of 10 µM. After two weeks of growth, colonies corresponding to each sample were counted. Each colony of C.reinhardtii represents a successful transformation of the resistance and indicates the expression of the aminoglycoside 3’-phosphotransferase. By counting the amount of colonies on the plates, we could determine which construct and at which DNA mass at the time of transformation worked best. | ||
| + | |||
| + | <h3>Improvement: Results </h3> | ||
| + | |||
| + | First, we counted the total number of <i>C. reinhardtii</i> colonies that were transformed with the standard and improved resistance, regardless of the DNA mass used at the time of transformation. The results of the total colony count can be seen in Fig. 1. Counting the colonies we discovered that the total number of colonies was much higher for the improved plasmid version. The colonies of the samples using the standard usage resulted in a total amount of 175 whereas the improved plasmid version produced 665 colonies. This result indicates that the improved paromomycin resistance works better than the one with standard codon usage. | ||
| + | |||
| + | <figure> | ||
| + | <img src="https://2019.igem.org/wiki/images/a/a7/T--Humboldt_Berlin--Colony_Amount_Total.png" alt="colonies_total" width="500"> | ||
| + | <figcaption>Fig.1 - Total amount of colonies counted for the standard (blue) and improved (organge) resistance</figcaption> | ||
| + | </figure> | ||
| + | |||
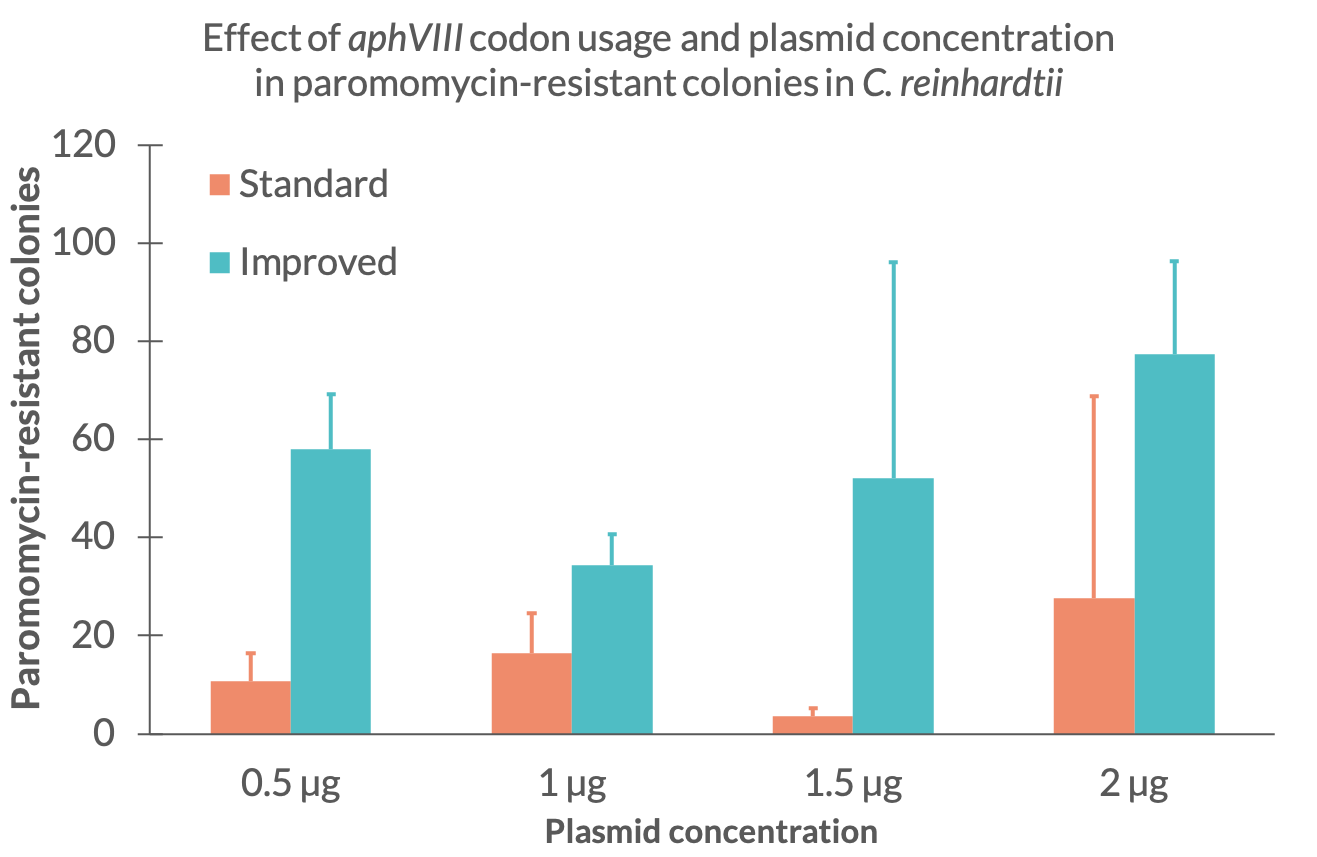
| + | Comparing the plasmid mass at the time of electroporation we discovered that the amount of colonies does not strictly correlate to the amount of DNA used during the electroporation (Fig. 2.). For the paromomycin resistance with standard codon usage we can see that the number of colonies at 1,5 µg is smaller than expected. Similarly, the amount of colonies for the improved resistance at a DNA mass of 0,5 µg is much higher than expected. The other results seem to show a tendency of increasing colony numbers with more DNA mass. Yet, further tests should be made to examine the exact effect of DNA mass during transformation for these parts. | ||
| + | |||
| + | <figure> | ||
| + | <img src="https://2019.igem.org/wiki/images/9/9d/T--Humboldt_Berlin--Colony_Amount_Mean_3.png" alt="colonies_mean" width="600"> | ||
| + | <figcaption>Fig 2. - Mean colony amount for the standard (blue) and improved (orange) paromomycin resistance for different DNA mass at the time of electroporation</figcaption> | ||
| + | </figure> | ||
| + | |||
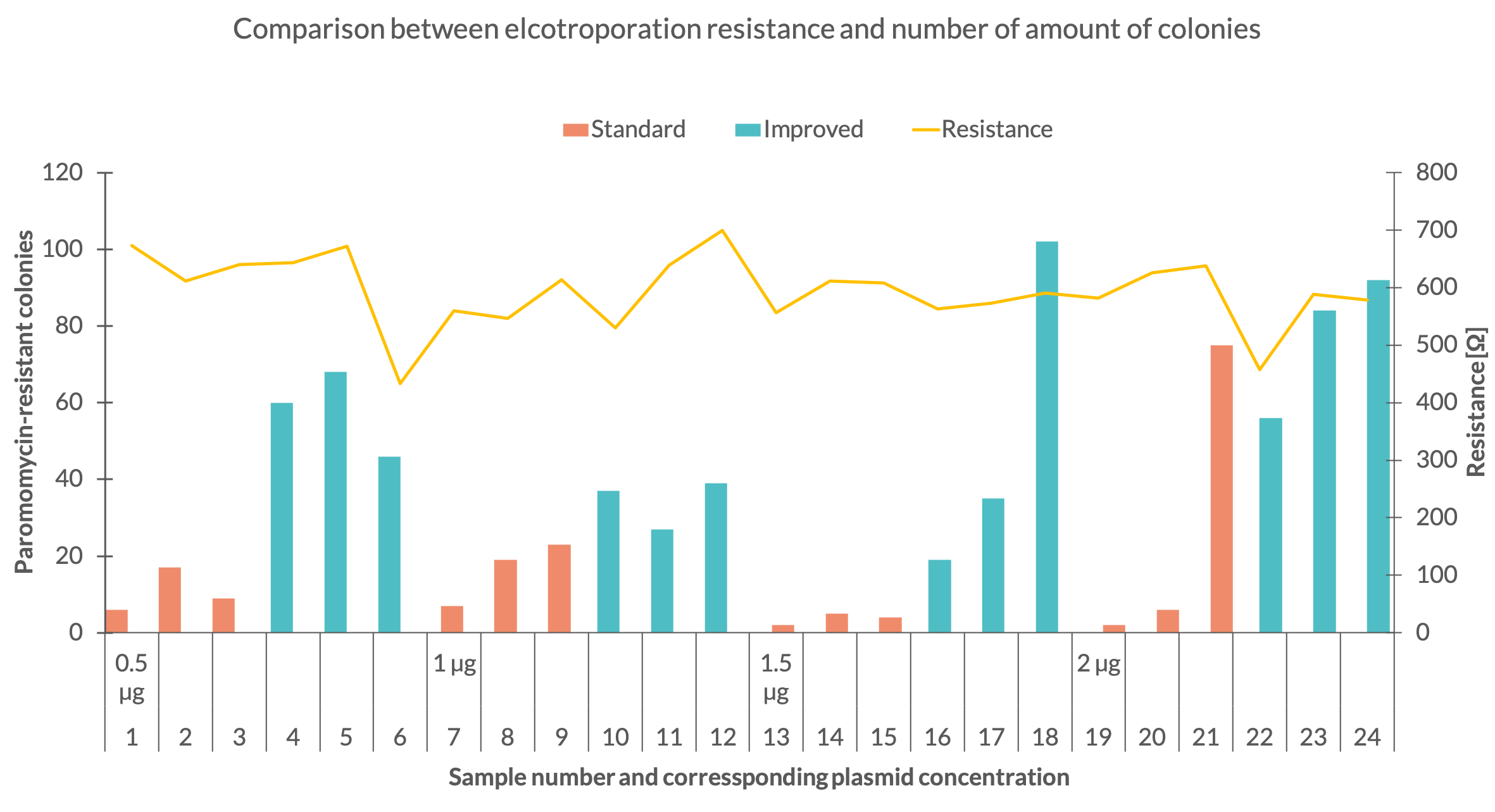
| + | One explanation for the variable amount of colonies might be the inconsistency of the electroporation resistance. To see how the electroporation process affected the number of colonies their quantity was compared with the corresponding resistance (Fig. 3). Fig. 3 depicts that the set with 1,5 µg standard plasmid was executed with a robust resistance of around 570 ohm for all 3 samples. The 1 µg set of the same plasmid shows a variable resistance but delivered more colonies. In the 2 µg standard plasmid set the resistance of the first sample dropped to 457 ohm but the same amount of colonies as in sample 1 of the 1,5 µg standard set were counted. With further comparison of these to sets it can be seen, that the third samples in the 1,5 µg and 2 µg sets showed similar resistance but the third sample of the second set resulted in a much higher colony number. The data behaves similar for the improved plasmid. The second sample of the first set and the third sample of the third were carried out with a resistance around 610 ohm but for the third sample almost 34 more colonies were counted. | ||
| + | |||
| + | <figure> | ||
| + | <img src="https://2019.igem.org/wiki/images/c/c8/T--Humboldt_Berlin--Colonies_with_resistance.png" alt="colonies_resistance" width="700"> | ||
| + | <figcaption>Fig 3. - Amount of colonies for each sample in dependency of the DNA mass and plotted with the electrical resistance of the electroporation</figcaption> | ||
| + | </figure> | ||
| + | |||
| + | The experiments showed clearly, that the improved codon usage resulted in larger number of <i>C.reinahrdtii</i> colonies capable of growing TAP-paromomycin-agar plates. This proves that the changed codon usage promotes a higher expression of the aminoglycoside 3’-phosphotransferase and is therefore an improvement of part BBa_K2703008. | ||
| + | |||
| + | </html> | ||
=='''References'''== | =='''References'''== | ||
| Line 88: | Line 128: | ||
<li> Weber, E., Engler, C., Gruetzner, R., Werner, S. & Marillonnet, S. A modular cloning system for standardized assembly of multigene constructs. PLoS One 6, (2011).</li> | <li> Weber, E., Engler, C., Gruetzner, R., Werner, S. & Marillonnet, S. A modular cloning system for standardized assembly of multigene constructs. PLoS One 6, (2011).</li> | ||
<li>Crozet, P. et al. Birth of a Photosynthetic Chassis: A MoClo Toolkit Enabling Synthetic Biology in the Microalga Chlamydomonas reinhardtii. ACS Synth. Biol. (2018). doi:10.1021/acssynbio.8b00251 </li> | <li>Crozet, P. et al. Birth of a Photosynthetic Chassis: A MoClo Toolkit Enabling Synthetic Biology in the Microalga Chlamydomonas reinhardtii. ACS Synth. Biol. (2018). doi:10.1021/acssynbio.8b00251 </li> | ||
| + | <li> Neupert, J., Karcher, D., & Bock, R. (2009). Generation of Chlamydomonas strains that efficiently express nuclear transgenes. The Plant Journal, 57(6), 1140-1150. | ||
| + | </li> | ||
</ol> | </ol> | ||
Latest revision as of 14:30, 13 October 2019
paromycin resistance gene
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Introduction
For the silver medal criteria “Validated Part” we submit a basic part: the sequence of resistance marker to paromomycine that is an aminoglycoside antibiotic. The aphVIII gene from Streptomyces rimosus encode for an aminoglycoside 3’-phosphotransferase gene (aph). From the aphVIII gene sequence of the BioBrick BBa_K1884013 (iGEM16_SZU-China) BpiI estriction site were removed ( G551C) by PCR point mutation. We cloned aphVIII by PCR amplification in B4-B5 position of the MoClo standard2 to use it in our retrotransposon design. Acceptor plasmid is the MoClo Universal pL0 acceptor plasmid pAGM91212. We sequenced again the sequence to be sure there is no unwanted mutations.
1- Biological background
It is usually use in Chlamydomonas reinhardtii under the control of the HSP70A promoter and the 3’UTR of RBCS2 gene as terminator1. Here we report the domestication of the aphVIII gene in the C. reinhardtii MoClo Kit
2- Usage in iGEM projects
Our part is compatible with the PhytoBrick MoClo standard. This PhytoBrick was intended to be used as a rapporter gene in the CARGO to test the functionality of our synthetic retrotransposons for directed evolution in C. reinhardtii.
Sequencing
Characterization
We couldn’t directly use our Phytobrick for its characterization because we did not have the time to construct all the other parts needed for the final functional assembly. Instead we used a slightly different Phytobrick with the same aphVIII sequence but with different fusion site in 5’ and 3’.
| Name | F1 : B4 | PART | F2: B5 |
| pL0-Paro | AGGT | aphVIII | TTCG |
This part was assembled into a functional Transcription Unit with. After transformation in C. reinhardtii D66 strain3 the functionality aphVIII gene sequence was tested by counting the number of colony resistant to paromomycine (15 ug/mL).
| Name | F1 | Prom | F2 | 5'UTR | F3 | Resistance | F4 | 3'UTR | F5 |
| pCM-1 | GGAG | PAR | TACT | 5’UTRAR | AATG | Paromycin | GCTT | TRBCS2 | CGCT |

Improvement
The Humboldt Team 2019 proposed an improvement of this part for the gold criterium #2. The improved part, BBa_K2984006, can be seen here. The part was improved by doing a codon optimization for use in Chlamydomonas reinhardtii.
Improvement: Methods
To see if the expression of the aminoglycoside 3’-phosphotransferase was increased, we performed several electroporations to transform C. reinhardtii with the improved paromomycin resistance. We used the C.reinhardtii strain UVM 4 since it is a strain designed to express transgene constructs (Neupert et al. 2009). We transformed the two paromomycin constructs with standard and improved codon usage starting with 0,5 µg DNA per electroporation sample and ascended with 0,5 µg steps up to 2 µg. For each construct and DNA mass we did three electroporations. The electroporation electrical resistance was measured for each sample. After resuspension and one day recovery in TAP medium, all samples were plated on TAP-agar plates containing a paromomycin concentration of 10 µM. After two weeks of growth, colonies corresponding to each sample were counted. Each colony of C.reinhardtii represents a successful transformation of the resistance and indicates the expression of the aminoglycoside 3’-phosphotransferase. By counting the amount of colonies on the plates, we could determine which construct and at which DNA mass at the time of transformation worked best.Improvement: Results
First, we counted the total number of C. reinhardtii colonies that were transformed with the standard and improved resistance, regardless of the DNA mass used at the time of transformation. The results of the total colony count can be seen in Fig. 1. Counting the colonies we discovered that the total number of colonies was much higher for the improved plasmid version. The colonies of the samples using the standard usage resulted in a total amount of 175 whereas the improved plasmid version produced 665 colonies. This result indicates that the improved paromomycin resistance works better than the one with standard codon usage.
References
- Sizova, I., Fuhrmann, M. & Hegemann, P. A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene 277, 221–229 (2001).
- Weber, E., Engler, C., Gruetzner, R., Werner, S. & Marillonnet, S. A modular cloning system for standardized assembly of multigene constructs. PLoS One 6, (2011).
- Crozet, P. et al. Birth of a Photosynthetic Chassis: A MoClo Toolkit Enabling Synthetic Biology in the Microalga Chlamydomonas reinhardtii. ACS Synth. Biol. (2018). doi:10.1021/acssynbio.8b00251
- Neupert, J., Karcher, D., & Bock, R. (2009). Generation of Chlamydomonas strains that efficiently express nuclear transgenes. The Plant Journal, 57(6), 1140-1150.
DOI: 10.1021/acssynbio.8b00251
