Difference between revisions of "Part:BBa K2669002:Experience"
Elinramstrom (Talk | contribs) |
m |
||
| (42 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| − | === | + | __NOTOC__ |
| + | ===Applications of BBa_K2669002 === | ||
| − | < | + | <br> |
| − | + | The construct [[BBa_K3189015]] containing the chromoprotein amilCP ([[BBa_K2669002]]) under the control of [[BBa_K3189001]]. When the system is induced with 100 ng/mL of tetracycline, a dark blue colour is produced (Figure 4 and Figure 5). This shows [[BBa_K3189001]] is able to function with different reporter proteins other than just <i>gfp</i>. | |
| − | < | + | <br> |
| − | + | <br> | |
| − | < | + | https://static.igem.org/mediawiki/parts/thumb/f/f5/T--Guelph--pTA-tetnotet.jpg/216px-T--Guelph--pTA-tetnotet.jpg |
| − | < | + | <br> |
| − | + | <b>Figure 1: [[BBa_K2669002]] under the control of [[BBa_K3189001]].</b> amilCP expression being induced using 100 ng/mL tetracycline (left) and uninduced (right). | |
| − | + | <br> | |
| − | + | <br> | |
| − | + | https://static.igem.org/mediawiki/parts/thumb/2/29/T--Guelph--pTAtetnotetspun.jpg/207px-T--Guelph--pTAtetnotetspun.jpg | |
| − | < | + | <br> |
| − | + | <b>Figure 2: Pellets of cells of [[BBa_K2669002]] under the control of [[BBa_K3189001]] induced and uninduced with tetracycline.</b> Pellet of cells induced with 100 ng/mL tetracycline (left) and pellet of cells uninduced (right). | |
| − | < | + | <br> |
| − | < | + | <br> |
| − | + | This year iGEM Guelph was working with BBa_K2669002 as a reporter protein. When working with this chromoprotein in <i>E.coli</i> BL21(DE3) under the control of [[BBa_K3189001]], we found that stronger colour was produced at lower temperatures compared to higher temperatures. In Figure 2 pigment intensity was compared between two temperatures, 25°C and 37°C. It is clear form these images that at 25°C the colonies have a darker colour compared to colonies grown at 37°C. This is evidence that AmilCP produced from [[BBa_K2669002]] is more stable at 25°C compared to 37°C thus allowing it to build up in the cells longer before degradation resulting in darker pigmentation of the colonies. | |
| − | + | <br> | |
| − | + | <br> | |
| − | + | https://2019.igem.org/wiki/images/thumb/f/f5/T--Guelph--AmilCPtemexpress.jpg/800px-T--Guelph--AmilCPtemexpress.jpg | |
| − | + | <br> | |
| − | + | <b>Figure 3: BBa_K2669002 expression at 25°C and 37°C.</b> Expression was carried out in <i>E.coli</i> BL21(DE3) on LB agar supplemented with 100 ug/mL ampicillin, 100 mg/mL tryptophan, 1mM IPTG, and 50 ng/mL tetracycline. | |
| − | + | <br> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | < | + | |
| − | < | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | < | + | |
| − | + | ||
| − | < | + | |
| − | + | ||
| − | + | ||
===User Reviews=== | ===User Reviews=== | ||
Latest revision as of 01:50, 22 October 2019
Applications of BBa_K2669002
The construct BBa_K3189015 containing the chromoprotein amilCP (BBa_K2669002) under the control of BBa_K3189001. When the system is induced with 100 ng/mL of tetracycline, a dark blue colour is produced (Figure 4 and Figure 5). This shows BBa_K3189001 is able to function with different reporter proteins other than just gfp.

Figure 1: BBa_K2669002 under the control of BBa_K3189001. amilCP expression being induced using 100 ng/mL tetracycline (left) and uninduced (right).

Figure 2: Pellets of cells of BBa_K2669002 under the control of BBa_K3189001 induced and uninduced with tetracycline. Pellet of cells induced with 100 ng/mL tetracycline (left) and pellet of cells uninduced (right).
This year iGEM Guelph was working with BBa_K2669002 as a reporter protein. When working with this chromoprotein in E.coli BL21(DE3) under the control of BBa_K3189001, we found that stronger colour was produced at lower temperatures compared to higher temperatures. In Figure 2 pigment intensity was compared between two temperatures, 25°C and 37°C. It is clear form these images that at 25°C the colonies have a darker colour compared to colonies grown at 37°C. This is evidence that AmilCP produced from BBa_K2669002 is more stable at 25°C compared to 37°C thus allowing it to build up in the cells longer before degradation resulting in darker pigmentation of the colonies.
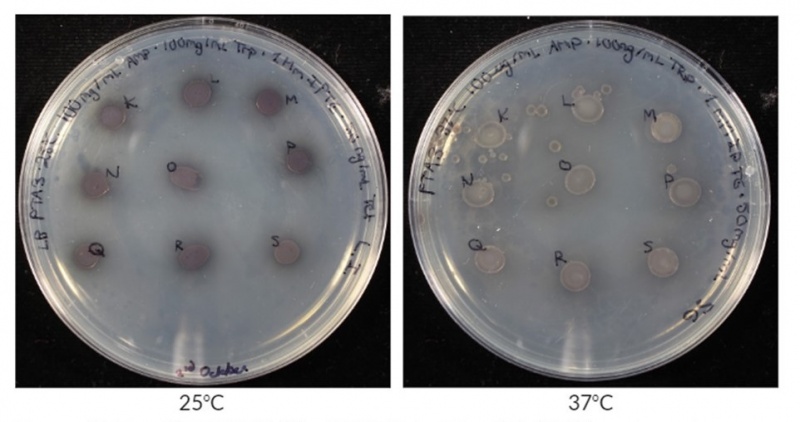
Figure 3: BBa_K2669002 expression at 25°C and 37°C. Expression was carried out in E.coli BL21(DE3) on LB agar supplemented with 100 ug/mL ampicillin, 100 mg/mL tryptophan, 1mM IPTG, and 50 ng/mL tetracycline.
User Reviews
UNIQ16e1a089a1109c9c-partinfo-00000000-QINU UNIQ16e1a089a1109c9c-partinfo-00000001-QINU
