Difference between revisions of "Part:BBa K2549013"
| (7 intermediate revisions by one other user not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K2549013 short</partinfo> | <partinfo>BBa_K2549013 short</partinfo> | ||
| − | This part is a mutant TEV(tobacco etch virus) protease. The | + | This part is a mutant TEV (tobacco etch virus) protease. The residue 219 of the wild-type TEV protease is mutated from serine to valine (S219V) to remove autoinactivation and perform a more efficient cleavage<ref>Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Kapust RB, Tözsér J, Fox JD, ..., Copeland TD, Waugh DS. Protein Eng, 2001 Dec;14(12):993-1000 PMID: 11809930</ref>. TEV protease is one of the best-characterized enzymes of the viral proteases which have more stringent sequence specificity. We used it to build our IMPLY logic gates. It can also be utilized by other iGEM teams to create their own engineered fusion proteins cleavage system. |
| − | |||
| − | |||
| − | |||
| − | |||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_K2549013 SequenceAndFeatures</partinfo> | <partinfo>BBa_K2549013 SequenceAndFeatures</partinfo> | ||
| − | === | + | <!-- Add more about the biology of this part here --> |
| − | + | ===Applications=== | |
| − | + | The main use of TEV protease<ref>https://en.wikipedia.org/wiki/TEV_protease</ref> is for removing affinity tags from purified recombinant fusion proteins. The reason for that is due to the high sequence specificity. This specificity allows for the controlled cleavage of proteins when the preference sequence is inserted into flexible loops. It also makes it relatively non-toxic in vivo as the recognized sequence scarcely occurs in proteins. | |
| − | + | ||
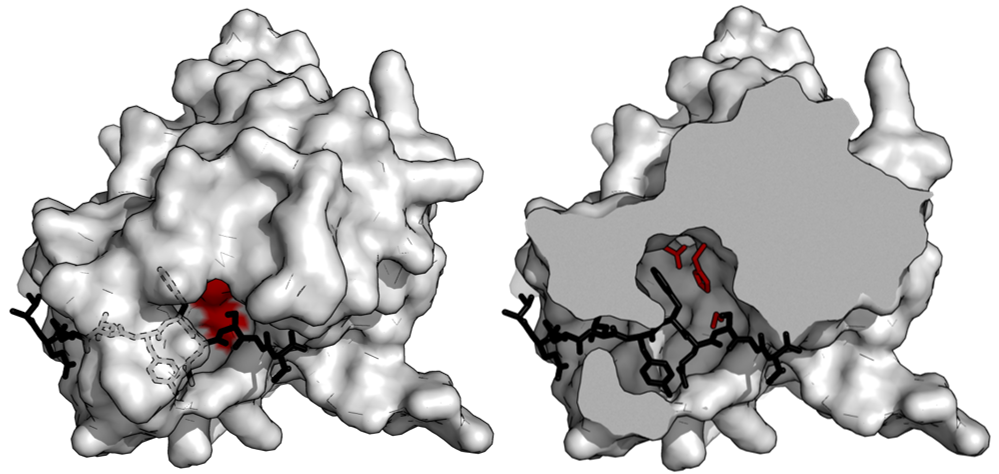
| + | [[File:TEV2.png|none|400px|thumb|wikipedia: ''Surface model of TEV bound to uncleaved substrate (black), also showing the catalytic triad (red). The substrate binds inside an active site tunnel (left). A cutaway (right) shows the complementary shape of the binding tunnel to the substrate. (PDB: 1lvb)'']] | ||
| + | |||
| + | [[File:TEV1.png|none|400px|thumb|Waugh DS wrote: ''TEV protease sequence specificity is far more stringent than that of factor Xa, thrombin, or enterokinase. It is a very useful reagent for cleaving fusion proteins. TEV protease recognizes a linear epitope of the general form E-Xaa-Xaa-Y-Xaa-Q-(G/S), with cleavage occurring between Q and G or Q and S.'']] | ||
| + | Please refer to [[Media:Tev-faq-7pages.pdf]] for more details on TEV protease. | ||
| + | |||
| + | ==== TEV in Tango Assay for GPCRs ==== | ||
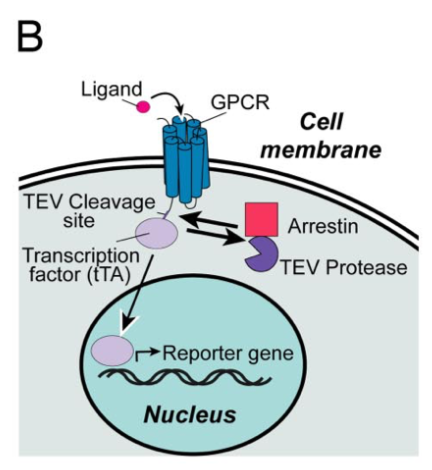
| + | Barnea G et al 2008<ref>The genetic design of signaling cascades to record receptor activation. Barnea G, Strapps W, Herrada G, ..., Axel R, Lee KJ. Proc Natl Acad Sci U S A, 2008 Jan;105(1):64-9 PMID: 18165312; DOI: 10.1073/pnas.0710487105</ref> wrote: ''We have developed an experimental strategy to monitor protein interactions in a cell with a high degree of selectivity and sensitivity. A transcription factor is tethered to a membrane-bound receptor with a linker that contains a cleavage site for a specific protease. Activation of the receptor recruits a signaling protein fused to the protease that then cleaves and releases the transcription factor to activate reporter genes in the nucleus. This strategy converts a transient interaction into a stable and amplifiable reporter gene signal to record the activation of a receptor without interference from endogenous signaling pathways. We have developed this assay for three classes of receptors: G protein-coupled receptors, receptor tyrosine kinases, and steroid hormone receptors. Finally, we use the assay to identify a ligand for the orphan receptor GPR1, suggesting a role for this receptor in the regulation of inflammation.'' | ||
| + | |||
| + | [[File:TEV3.png|none|200px|thumb|Barnea G et al stated: ''Schematic of the Tango assay method to monitor GPCR–arrestin interactions. Ligand binding to the target receptor stimulates recruitment of the arrestin-TEV protease fusion, triggering the release of the tethered transcription factor tTA. Free tTA then enters the nucleus and stimulates reporter gene activity.'']] | ||
| + | |||
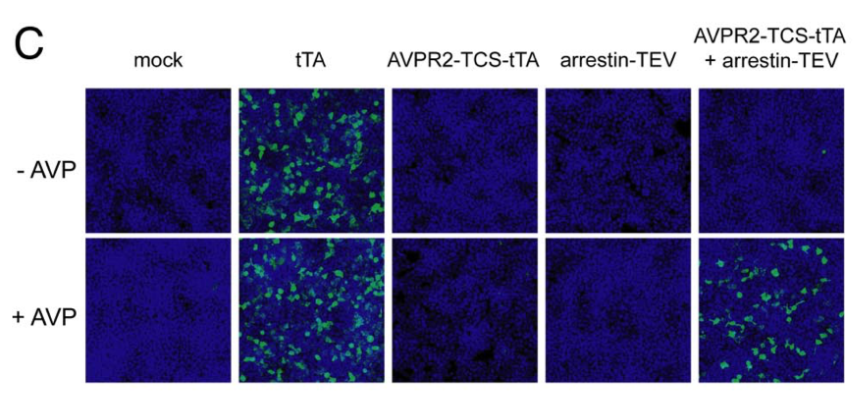
| + | [[File:TEV4.png|none|400px|thumb|Barnea G et al stated: ''Tango assay for the arginine vasopressin receptor AVPR2. HTL cells (an HEK293T-derived cell line containing a stably integrated tTA-dependent luciferase reporter) were transiently transfected with expression plasmids encoding the arginine vasopressin receptor 2 (AVPR2)-TEV cleavage site-tTA (AVPR2-TCS-tTA) and β-arrestin2-TEV protease (Arrestin-TEV) fusion proteins, as indicated. Control cells were transfected either with a tTA expression plasmid (tTA) or with empty parental expression plasmid (mock). Luciferase expression was detected by immunostaining with anti-luciferase (green), and cell nuclei were visualized with the DNA dye TOTO-3 (blue). Western blot analysis confirmed that receptor and arrestin components were expressed at similar levels in each transfection.'']] | ||
| + | |||
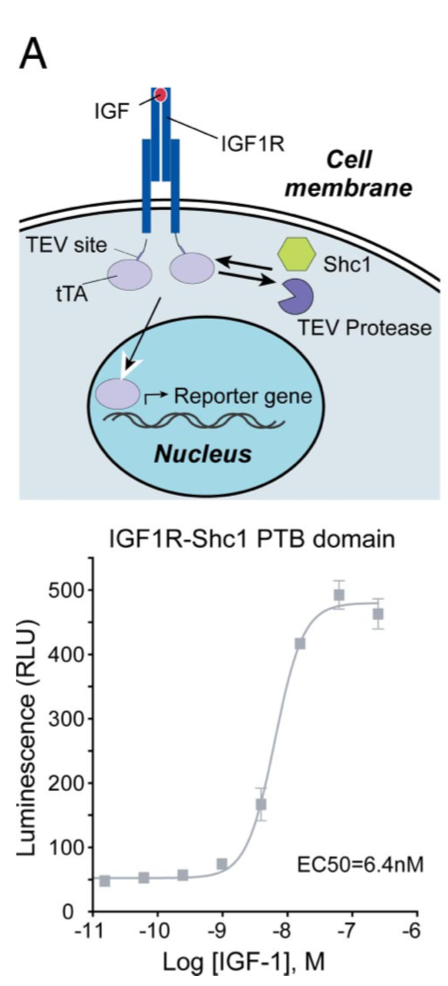
| + | [[File:TEV5.png|none|200px|thumb|Barnea G et al stated: ''Receptor tyrosine kinase signaling. HTL cells were transfected with an IGF-1 receptor (IGF1R)–TCS-tTA fusion construct and a Shc1 PTB domain-TEV protease fusion plasmid. Luciferase activity in this IGF1R–Shc1 interaction assay was stimulated by IGF-1. All error bars represent SD (n = four measurements).'']] | ||
| + | |||
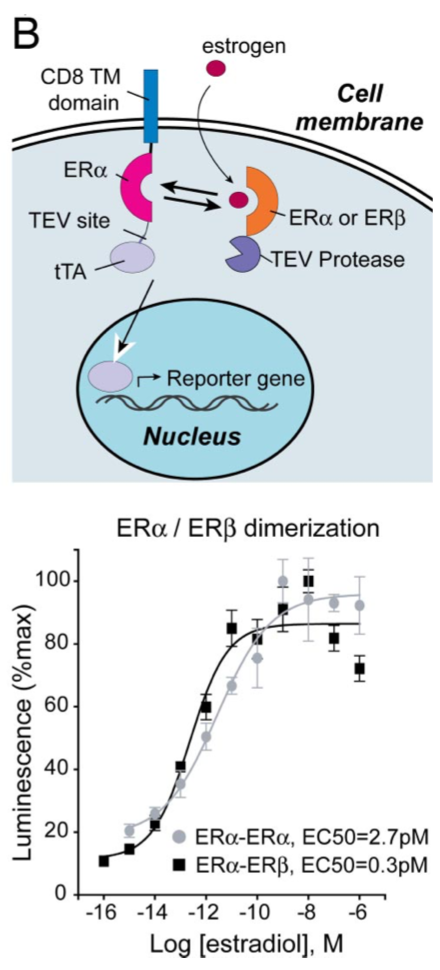
| + | [[File:TEV6.png|none|200px|thumb|Barnea G et al stated: ''Ligand-induced homo- and heterodimerization of estrogen receptors (ERs). HTL cells were transfected with a CD8-ERα-TCS-tTA fusion and either an ERα- or ERβ-TEV protease fusion to measure ERα homodimerization (light circles) or ERα/ERβ heterodimerization (dark squares) as illustrated. Responses were normalized to the maximal response for each receptor. All error bars represent SD (n = four measurements).'']] | ||
| + | |||
| + | ==== TEV in ChaCha Design ==== | ||
| + | Kipniss NH et al 2017<ref>Engineering cell sensing and responses using a GPCR-coupled CRISPR-Cas system. Kipniss NH, Dingal PCDP, Abbott TR, ..., Labanieh L, Qi LS. Nat Commun, 2017 Dec;8(1):2212 PMID: 29263378; DOI: 10.1038/s41467-017-02075-1</ref> wrote: ''We found that the hM3D-CRISPR ChaCha exhibited better reporter gene activation (4.4 fold) compared to the hM3D- CRISPR Tango (1.2 fold) after 1-day treatment with CNO. The Tango design displayed higher leakiness than the ChaCha design with significant GFP expression without CNO treatment. The observation that ChaCha outperforms Tango may suggest that fusing the small TEVp (28 kDa) to the receptor likely preserves the conformational fidelity and activity of the GPCR molecule, while fusing a larger domain such as dCas9-VPR (220 kDa) to GPCR may compromise its conformation by exposing the C-terminal tail of a GPCR and permit leaky interactions with ARRB2.'' | ||
| + | |||
| + | [[File:TEV7.png|none|500px|thumb|Barnea G et al stated: ''(a) Two design schemes of coupling CRISPR-Cas9 function to the activity of GPCR. '''Left:''' the Tango design fused the effector protein (dCas9-VPR as shown) to the C-terminus of GPCR via a V2 tail sequence and a TEV cleavage sequence (TCS). An adaptor protein, β-Arrestin 2 (ARRB2), is fused to the Tobacco Etch Virus protease (TEVp). '''Right:''' in the ChaCha design, we fused the effector to the adaptor protein via the TCS, and fused the TEVp to the receptor via the V2 tail. Upon ligand binding to the receptor, both systems recruit the adaptor to the V2 tail, and the protease specifically cleaves at the TCS, releasing the effector protein that translocates into the nucleus for sgRNA-directed gene regulation.<br/>(b) Comparison of the performance of Tango (orange) and ChaCha (blue) designs using the synthetically evolved GPCR, hM3D in HEK293T cells with or without 20 μM clozapine-N-oxide (CNO), the ligand for hM3D (see Methods section for detailed experimental procedure). The free dCas9-VPR (gray) with and without a targeting sgRNA (sgTet) are used as positive and negative controls. The data were normalized to the free dCas9-VPR without a targeting sgRNA. The data represent two independent experiments with 4–8 technical replicates, and the bars represent the mean.'']] | ||
| + | |||
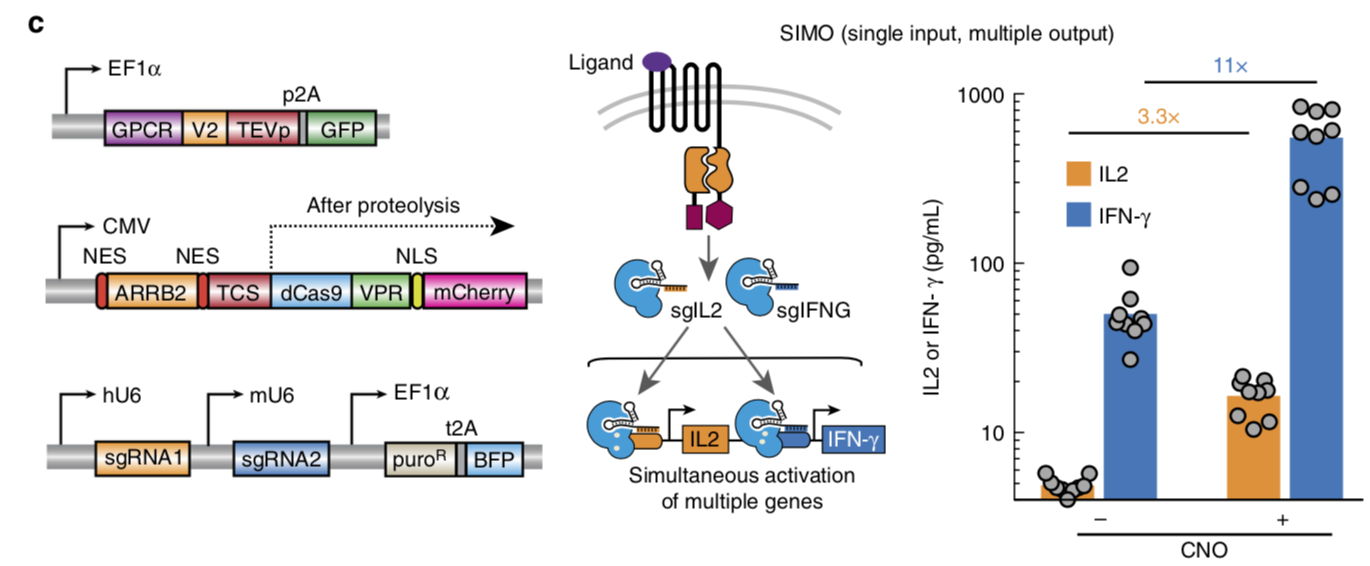
| + | [[File:TEV8.png|none|500px|thumb|Barnea G et al stated: ''Simultaneous activation of two endogenous genes using the hM3D-CRISPR ChaCha system. '''Left,''' the plasmids of ChaCha system used for multiplexed gene regulation. '''Middle,''' a schematic overview of the ChaCha SIMO (single input, multiple output) system. '''Right,''' ELISA measure of both endogenous IL2 (orange) and IFN-γ (blue) expression and secretion using the ChaCha SIMO. For ELISA measurements, bars are the mean of three independent experiments with 7–9 technical replicates. For qPCR measurements, bars are the mean of three independent experiments that contained three technical replicates, which are then measured in technical qPCR duplicate. See Methods section for detailed experimental procedure'']] | ||
| + | |||
| + | |||
| + | Note: [[Part:BBa_K2549013]], [[Part:BBa_K2549014]] and [[Part:BBa_K2549015]] have the same Applications section. | ||
| Line 22: | Line 42: | ||
===Functional Parameters=== | ===Functional Parameters=== | ||
<partinfo>BBa_K2549013 parameters</partinfo> | <partinfo>BBa_K2549013 parameters</partinfo> | ||
| − | + | --> | |
| + | |||
| + | ===References=== | ||
Latest revision as of 03:18, 12 October 2018
TEV protease
This part is a mutant TEV (tobacco etch virus) protease. The residue 219 of the wild-type TEV protease is mutated from serine to valine (S219V) to remove autoinactivation and perform a more efficient cleavage[1]. TEV protease is one of the best-characterized enzymes of the viral proteases which have more stringent sequence specificity. We used it to build our IMPLY logic gates. It can also be utilized by other iGEM teams to create their own engineered fusion proteins cleavage system.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 316
Applications
The main use of TEV protease[2] is for removing affinity tags from purified recombinant fusion proteins. The reason for that is due to the high sequence specificity. This specificity allows for the controlled cleavage of proteins when the preference sequence is inserted into flexible loops. It also makes it relatively non-toxic in vivo as the recognized sequence scarcely occurs in proteins.
Please refer to Media:Tev-faq-7pages.pdf for more details on TEV protease.
TEV in Tango Assay for GPCRs
Barnea G et al 2008[3] wrote: We have developed an experimental strategy to monitor protein interactions in a cell with a high degree of selectivity and sensitivity. A transcription factor is tethered to a membrane-bound receptor with a linker that contains a cleavage site for a specific protease. Activation of the receptor recruits a signaling protein fused to the protease that then cleaves and releases the transcription factor to activate reporter genes in the nucleus. This strategy converts a transient interaction into a stable and amplifiable reporter gene signal to record the activation of a receptor without interference from endogenous signaling pathways. We have developed this assay for three classes of receptors: G protein-coupled receptors, receptor tyrosine kinases, and steroid hormone receptors. Finally, we use the assay to identify a ligand for the orphan receptor GPR1, suggesting a role for this receptor in the regulation of inflammation.
TEV in ChaCha Design
Kipniss NH et al 2017[4] wrote: We found that the hM3D-CRISPR ChaCha exhibited better reporter gene activation (4.4 fold) compared to the hM3D- CRISPR Tango (1.2 fold) after 1-day treatment with CNO. The Tango design displayed higher leakiness than the ChaCha design with significant GFP expression without CNO treatment. The observation that ChaCha outperforms Tango may suggest that fusing the small TEVp (28 kDa) to the receptor likely preserves the conformational fidelity and activity of the GPCR molecule, while fusing a larger domain such as dCas9-VPR (220 kDa) to GPCR may compromise its conformation by exposing the C-terminal tail of a GPCR and permit leaky interactions with ARRB2.
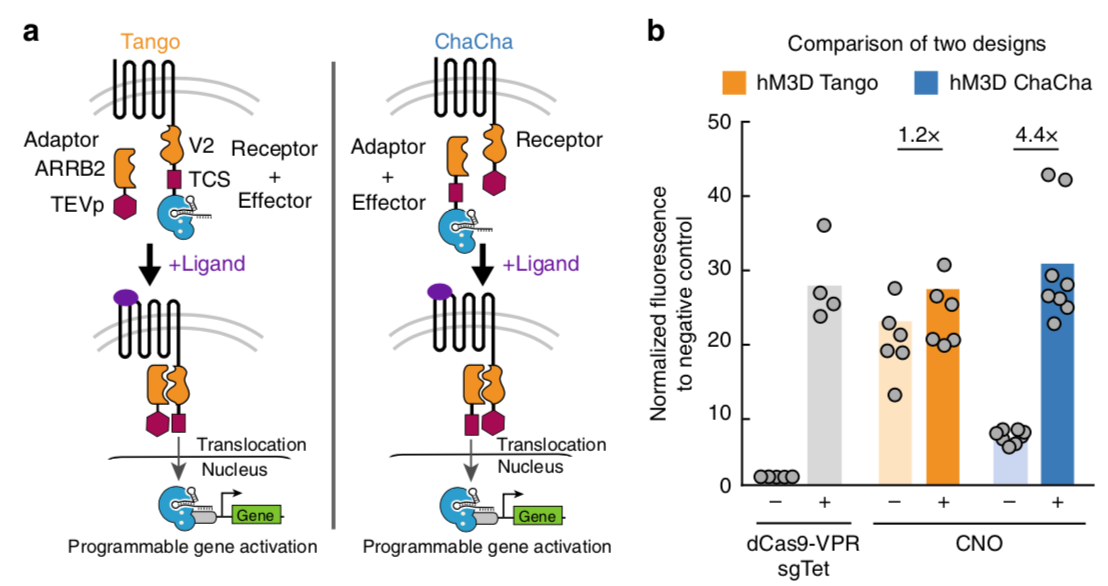
(b) Comparison of the performance of Tango (orange) and ChaCha (blue) designs using the synthetically evolved GPCR, hM3D in HEK293T cells with or without 20 μM clozapine-N-oxide (CNO), the ligand for hM3D (see Methods section for detailed experimental procedure). The free dCas9-VPR (gray) with and without a targeting sgRNA (sgTet) are used as positive and negative controls. The data were normalized to the free dCas9-VPR without a targeting sgRNA. The data represent two independent experiments with 4–8 technical replicates, and the bars represent the mean.
Note: Part:BBa_K2549013, Part:BBa_K2549014 and Part:BBa_K2549015 have the same Applications section.
References
- ↑ Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Kapust RB, Tözsér J, Fox JD, ..., Copeland TD, Waugh DS. Protein Eng, 2001 Dec;14(12):993-1000 PMID: 11809930
- ↑ https://en.wikipedia.org/wiki/TEV_protease
- ↑ The genetic design of signaling cascades to record receptor activation. Barnea G, Strapps W, Herrada G, ..., Axel R, Lee KJ. Proc Natl Acad Sci U S A, 2008 Jan;105(1):64-9 PMID: 18165312; DOI: 10.1073/pnas.0710487105
- ↑ Engineering cell sensing and responses using a GPCR-coupled CRISPR-Cas system. Kipniss NH, Dingal PCDP, Abbott TR, ..., Labanieh L, Qi LS. Nat Commun, 2017 Dec;8(1):2212 PMID: 29263378; DOI: 10.1038/s41467-017-02075-1