Difference between revisions of "Part:BBa K2442101"
(→Part Improvement) |
|||
| (8 intermediate revisions by 4 users not shown) | |||
| Line 4: | Line 4: | ||
<p>This part contains pBAD promoter on its own. pBAD promoter is derived from the L-arabinose operon from ''Escherichia coli''. This version of pBAD retains all binding sites for the regulatory protein AraC, but lacks the full araC coding region. Potential "araC" start codon has been mutated from ATG to AGT.</p> | <p>This part contains pBAD promoter on its own. pBAD promoter is derived from the L-arabinose operon from ''Escherichia coli''. This version of pBAD retains all binding sites for the regulatory protein AraC, but lacks the full araC coding region. Potential "araC" start codon has been mutated from ATG to AGT.</p> | ||
| − | <p>This promoter gives 2000x fold induction when on pSB1C3 backbone, or x300 fold induction when on low copy number plasmid pSB3k3 when used with BioBrick [https://parts.igem.org/Part:BBa_K2442104 BBa_K2442104] in a strain lacking | + | <p>This promoter gives 2000x fold induction when on pSB1C3 backbone, or x300 fold induction when on low copy number plasmid pSB3k3 when used with BioBrick [https://parts.igem.org/Part:BBa_K2442104 BBa_K2442104] in a strain lacking AraC.</p> |
<p>This BioBrick together with [https://parts.igem.org/Part:BBa_K2442104 BBa_K2442104] is an improvement of [https://parts.igem.org/Part:BBa_I0500 BBa_I0500] by splitting its constituent parts - pBAD promoter and ''araC'' - into two separate plasmids to allow greater control over production of AraC. </p> | <p>This BioBrick together with [https://parts.igem.org/Part:BBa_K2442104 BBa_K2442104] is an improvement of [https://parts.igem.org/Part:BBa_I0500 BBa_I0500] by splitting its constituent parts - pBAD promoter and ''araC'' - into two separate plasmids to allow greater control over production of AraC. </p> | ||
| Line 23: | Line 23: | ||
<p>In absence of L-arabinose the AraC dimer binds to operator half-sites O<sub>2</sub> and I<sub>1</sub>. This causes DNA looping upstream of the pBAD promoter which represses transcription by excluding RNA polymerase from binding to pBAD or PC. Binding of L-arabinose causes a conformational change in the protein such that the DNA-binding domains of the dimer bind to adjacent I<sub>1</sub> and I<sub>2</sub> half-sites, giving access for RNA polymerase and cyclic AMP receptor protein (CRP) to bind pBAD. This results in transcription activation of downstream genes.</p> | <p>In absence of L-arabinose the AraC dimer binds to operator half-sites O<sub>2</sub> and I<sub>1</sub>. This causes DNA looping upstream of the pBAD promoter which represses transcription by excluding RNA polymerase from binding to pBAD or PC. Binding of L-arabinose causes a conformational change in the protein such that the DNA-binding domains of the dimer bind to adjacent I<sub>1</sub> and I<sub>2</sub> half-sites, giving access for RNA polymerase and cyclic AMP receptor protein (CRP) to bind pBAD. This results in transcription activation of downstream genes.</p> | ||
| − | [[Image:T-Glasgow-K2442102- | + | [[Image:T-Glasgow-K2442102-Fig1c.png|centre|thumb|800px|'''Figure 1: Annotated sequence of minimal P<sub>BAD</sub>. AraC potential start codon mutation ATG→AGT position is highlighted in <font color="magenta">magenta</font> (in reverse compliment). AraC-binding half sites are underlined and annotated. -35 and -10 elements of the P<sub>BAD</sub> promoter are highlighted in <font color="cyan">cyan</font>. AraC transcription start site (+1) highlighted in <font color="grey">grey</font>. P<sub>C</sub> promoter and its direction is indicated by an <font color="orange">orange</font> arrow. -35 and -10 elements of the P<sub>C</sub> promoter are indicated.''']] |
<p>The aim of our project was to mutagenize 4 residues within the AraC ligand-binding pocket to generate mutants with altered effector specificity. To allow mutant screening we transformed AraC-negative ''E. coli'' strain, DH5α, with regulatory plasmid carrying the ''araC'' mutant library and reporter plasmid [https://parts.igem.org/Part:BBa_K2442102 BBa_K2442102] with pBAD and GFP. Mutants were tested for responsiveness to xylose, decanal, arabinose and no inducer based on fluorescence.</p> | <p>The aim of our project was to mutagenize 4 residues within the AraC ligand-binding pocket to generate mutants with altered effector specificity. To allow mutant screening we transformed AraC-negative ''E. coli'' strain, DH5α, with regulatory plasmid carrying the ''araC'' mutant library and reporter plasmid [https://parts.igem.org/Part:BBa_K2442102 BBa_K2442102] with pBAD and GFP. Mutants were tested for responsiveness to xylose, decanal, arabinose and no inducer based on fluorescence.</p> | ||
| Line 42: | Line 42: | ||
[[Image:T-Glasgow-I0500-Graph.png|centre|thumb|800px|'''Figure 3:Average relative fluorescence over optical density at hour 8. Shows fluorescence levels under: no additive, 40mM Arabinose, 40mM Xylose and 2mM decanal. The ''E. coli'' strains which the plasmids have been transformed into are DS941 or DH5α (specified). DH5α is AraC-positive. DS941 is AraC-negative. BioBricks (K244210..) specified. All parts are in plasmid backbone pSB1C3 unless specified otherwise (pSB3k3). K2442102: reporter plasmid pBADmin+GFP. K2442104: regulatory plasmid R0011+B0032+AraC''']] | [[Image:T-Glasgow-I0500-Graph.png|centre|thumb|800px|'''Figure 3:Average relative fluorescence over optical density at hour 8. Shows fluorescence levels under: no additive, 40mM Arabinose, 40mM Xylose and 2mM decanal. The ''E. coli'' strains which the plasmids have been transformed into are DS941 or DH5α (specified). DH5α is AraC-positive. DS941 is AraC-negative. BioBricks (K244210..) specified. All parts are in plasmid backbone pSB1C3 unless specified otherwise (pSB3k3). K2442102: reporter plasmid pBADmin+GFP. K2442104: regulatory plasmid R0011+B0032+AraC''']] | ||
| + | |||
| + | |||
| + | |||
| + | ===Contribution=== | ||
| + | Group: Valencia_UPV iGEM 2018 | ||
| + | <br> | ||
| + | Author: Adrián Requena Gutiérrez, Carolina Ropero | ||
| + | <br> | ||
| + | Summary: We adapted the part to be able to assemble transcriptional units with the Golden Gate assembly method | ||
| + | <br> | ||
| + | Documentation: | ||
| + | In order to create our complete [http://2018.igem.org/Team:Valencia_UPV/Part_Collection Printeria Part collection] of parts compatible with the Golden Gate assembly method, we made the part [https://parts.igem.org/Part:BBa_K2656006 BBa_K2656006] which is this part adapted to the Golden Gate technology. | ||
| + | |||
| + | === Part Improvement=== | ||
| + | In 2022, the team at Cambridge has designed a collection of nine pBAD promoters that are all only less than 160 base-pairs - half of the wild-type BBa_K2442101's length. Four members showed certain level of improvements, with two of those (called pBAD_AP2 and pBAD_AP3) showcased very promising merits, with slightly lower leakiness but more than fourfold increase in maximal expression. Please check out their documentation entry on [https://parts.igem.org/Part:BBa_K4491007 BBa_K4491007]. Below is a brief recap of the testing result. Also check out [https://parts.igem.org/Part:BBa_K2442101:Experience experience page] for brief result recap. | ||
| Line 47: | Line 62: | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_K2442101 SequenceAndFeatures</partinfo> | <partinfo>BBa_K2442101 SequenceAndFeatures</partinfo> | ||
| − | |||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
Latest revision as of 01:49, 14 October 2022
Minimal pBAD promoter
This part contains pBAD promoter on its own. pBAD promoter is derived from the L-arabinose operon from Escherichia coli. This version of pBAD retains all binding sites for the regulatory protein AraC, but lacks the full araC coding region. Potential "araC" start codon has been mutated from ATG to AGT.
This promoter gives 2000x fold induction when on pSB1C3 backbone, or x300 fold induction when on low copy number plasmid pSB3k3 when used with BioBrick BBa_K2442104 in a strain lacking AraC.
This BioBrick together with BBa_K2442104 is an improvement of BBa_I0500 by splitting its constituent parts - pBAD promoter and araC - into two separate plasmids to allow greater control over production of AraC.
This part belongs to a collection of parts made by [http://2017.igem.org/Team:Glasgow Team Glasgow 2017]:
- BBa_K2442101: minimal pBAD promoter
- BBa_K2442102: reporter plasmid - minimal pBAD promoter + GFP
- BBa_K2442103: araC coding sequence
- BBa_K2442104: regulatory plasmid - araC coding sequence under control of LacI-regulated promoter
Usage and Biology
The L-arabinose operon is naturally found in Escherichia coli. The regulatory protein, AraC, acts as a dimer. The operon contains two regulatory cis elements: the PC promoter for the synthesis of AraC, and the pBAD promoter for synthesis of enzymes required for catabolism of L-arabinose. pBAD promoter contains 3 half sites that each bind to one subunit of AraC - O2, I1, I2. The O1 site is composed of O1L and O1R half sites, which bind both subunits. The O2 half site is within the araC coding region.
To improve BBa_I0500, we split the araC and pBAD into two separate parts. We isolated the minimal DNA sequence from the native pBAD that is sufficient to retain its function. The promoter retains all the operator sites O1, O2, I1 and I2 required for binding of AraC (Fig. 1). As some of these sites lie within the PC promoter and overlap with AraC coding sequence, the minimal pBAD has the potential araC start codon ATG mutated to AGT to prevent the protein from being made.
In absence of L-arabinose the AraC dimer binds to operator half-sites O2 and I1. This causes DNA looping upstream of the pBAD promoter which represses transcription by excluding RNA polymerase from binding to pBAD or PC. Binding of L-arabinose causes a conformational change in the protein such that the DNA-binding domains of the dimer bind to adjacent I1 and I2 half-sites, giving access for RNA polymerase and cyclic AMP receptor protein (CRP) to bind pBAD. This results in transcription activation of downstream genes.
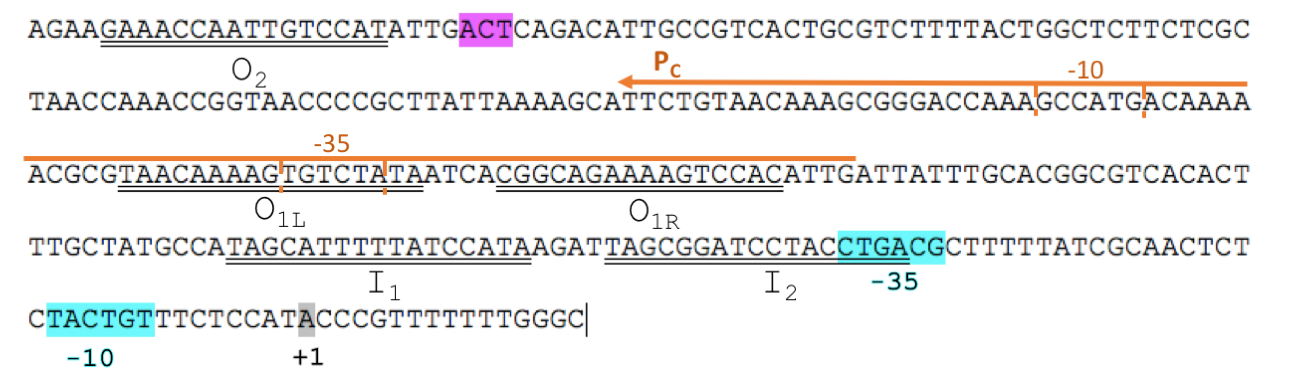
The aim of our project was to mutagenize 4 residues within the AraC ligand-binding pocket to generate mutants with altered effector specificity. To allow mutant screening we transformed AraC-negative E. coli strain, DH5α, with regulatory plasmid carrying the araC mutant library and reporter plasmid BBa_K2442102 with pBAD and GFP. Mutants were tested for responsiveness to xylose, decanal, arabinose and no inducer based on fluorescence.
pBAD promoter can be placed upstream of any gene of choice to construct arabinose -inducible systems. Moreover, our protocol for AraC mutagenesis can be followed to generate biosensors responsive to any other small molecule of interest, with BBa_K2442102 serving the role of reporter in mutant screens and detection systems. We generated 4 mutants which deviate in behaviour from wild-type araC: BBa_K2442105, BBa_K2442106, BBa_K2442107 and BBa_K2442108. Visit our [http://2017.igem.org/Team:Glasgow/araC wiki] for details.
Characterization
First of all, to test for the functionality of pBAD promoter we transformed BBa_K2442102 into E. coli strain DH5α. DH5α possesses a functional copy of arac on its chromosome. Transformants were restreaked onto LB agar with 0.2% arabinose, and 0.5% glucose as control (Fig. 2). 13 out of 14 picked transformants exhibited fluorescence under +arabinose conditions, and none of the colonies showed fluorescence under +glucose conditions. We therefore concluded that minimal pBAD is fully functional and is only induced in the presence of L-arabinose. Chromosomal copy of ‘’araC’’ showed to be sufficient to induce expression of GFP.

We then tested the part in presence of arabinose, xylose, decanal and no inducer (Fig. 3), as these were the compounds we used in mutant screening. Empty DH5α cells served as control for basal level of fluorescence. Supporting result obtained in previous experiment (Fig. 2), DH5α cells carrying BBa_K2442102 showed 2000x fold increase in fluorescence in presence of arabinose, but no significant increase in fluorescence on other sugars.
DH5α cells carrying minimal pBAD and I13500 on a low copy number plasmid pSB3K3 instead of pSB1C3 also showed expression of GFP only under arabinose conditions. However, levels of fluorescence were much lower, suggesting that the level of GFP production is dependent on the plasmid copy number and must be taken into account in further experiments.
We then transformed BBa_K2442102 into E. coli strain DS941, which does not possess a functional copy of araC on its chromosome. Empty DS941 cells served as control for basal level of fluorescence. Transformed cells showed no increase in fluorescence. This showed that AraC is necessary for activation of pBAD.
Next, we transformed DS941 cells with minimal pBAD+GFP in pSB3k3 and our regulatory plasmid encoding for AraC - BBa_K2442104 (pSB1C3). Increase in fluorescence was comparable to DH5α carrying minimal pBAD+GFP in pSB3k3 only. This shows that AraC expressed from BBa_K2442104 plasmid is sufficient to activate pBAD. Expressing AraC from a high copy number plasmid (pSB1C3) versus from a single chromosomal copy does not increase GFP expression.
Overall, we conclude that both L-arabinose and AraC are necessary to activate pBAD. pBAD promoter shows no leakage. 2000x fold increase in fluorescence of DH5α cells carrying BBa_K2442102 on arabinose shows high efficiency of pBAD and proves improvement over BBa_I0500

Contribution
Group: Valencia_UPV iGEM 2018
Author: Adrián Requena Gutiérrez, Carolina Ropero
Summary: We adapted the part to be able to assemble transcriptional units with the Golden Gate assembly method
Documentation:
In order to create our complete [http://2018.igem.org/Team:Valencia_UPV/Part_Collection Printeria Part collection] of parts compatible with the Golden Gate assembly method, we made the part BBa_K2656006 which is this part adapted to the Golden Gate technology.
Part Improvement
In 2022, the team at Cambridge has designed a collection of nine pBAD promoters that are all only less than 160 base-pairs - half of the wild-type BBa_K2442101's length. Four members showed certain level of improvements, with two of those (called pBAD_AP2 and pBAD_AP3) showcased very promising merits, with slightly lower leakiness but more than fourfold increase in maximal expression. Please check out their documentation entry on BBa_K4491007. Below is a brief recap of the testing result. Also check out experience page for brief result recap.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 241
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 76
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 58
