|
|
| (3 intermediate revisions by the same user not shown) |
| Line 2: |
Line 2: |
| | This experience page is provided so that any user may enter their experience using this part.<BR>Please enter | | This experience page is provided so that any user may enter their experience using this part.<BR>Please enter |
| | how you used this part and how it worked out. | | how you used this part and how it worked out. |
| − |
| |
| − | ===Characterization of BBa_C0060 by Tec-Chihuahua 2017===
| |
| − |
| |
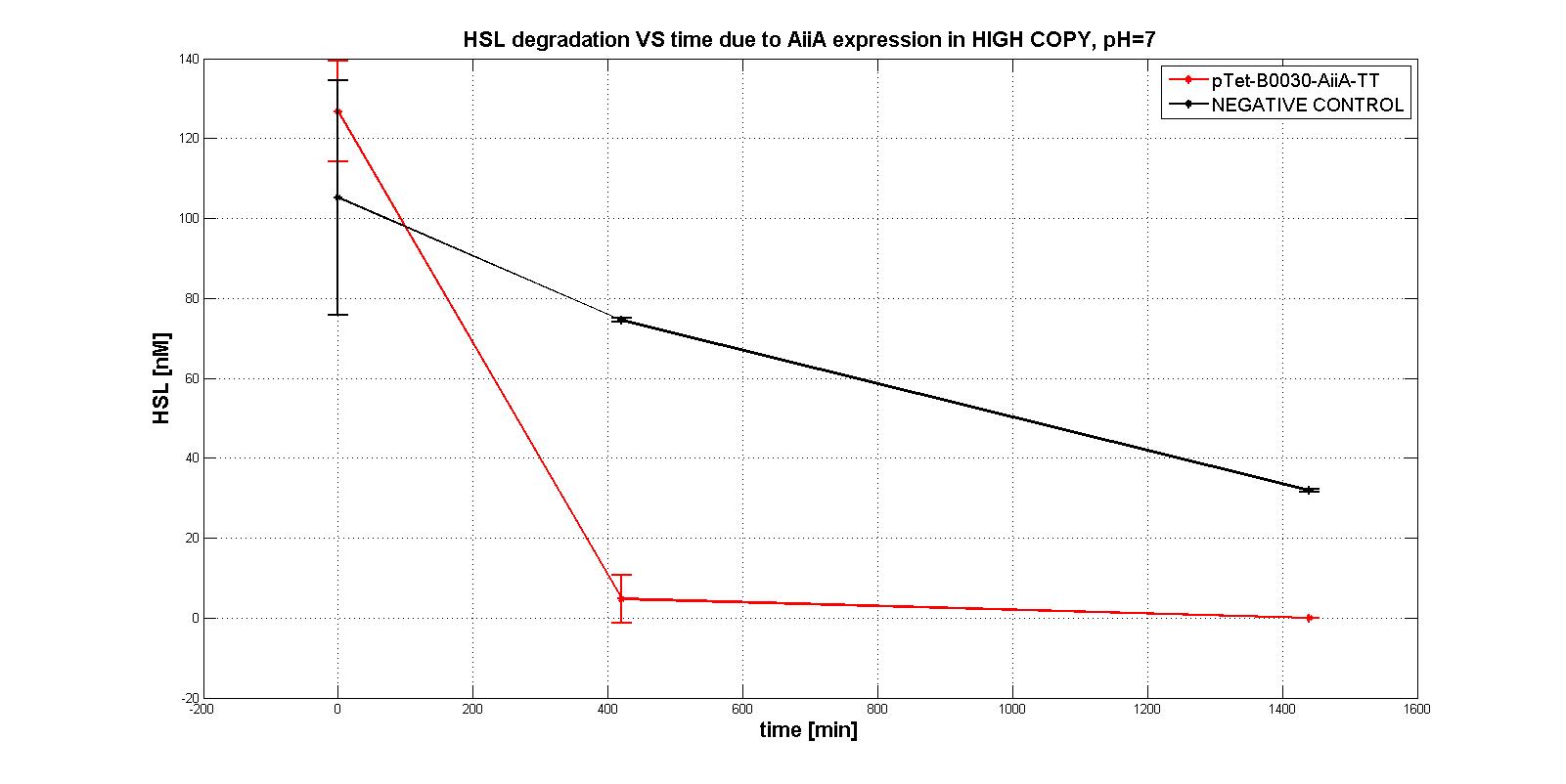
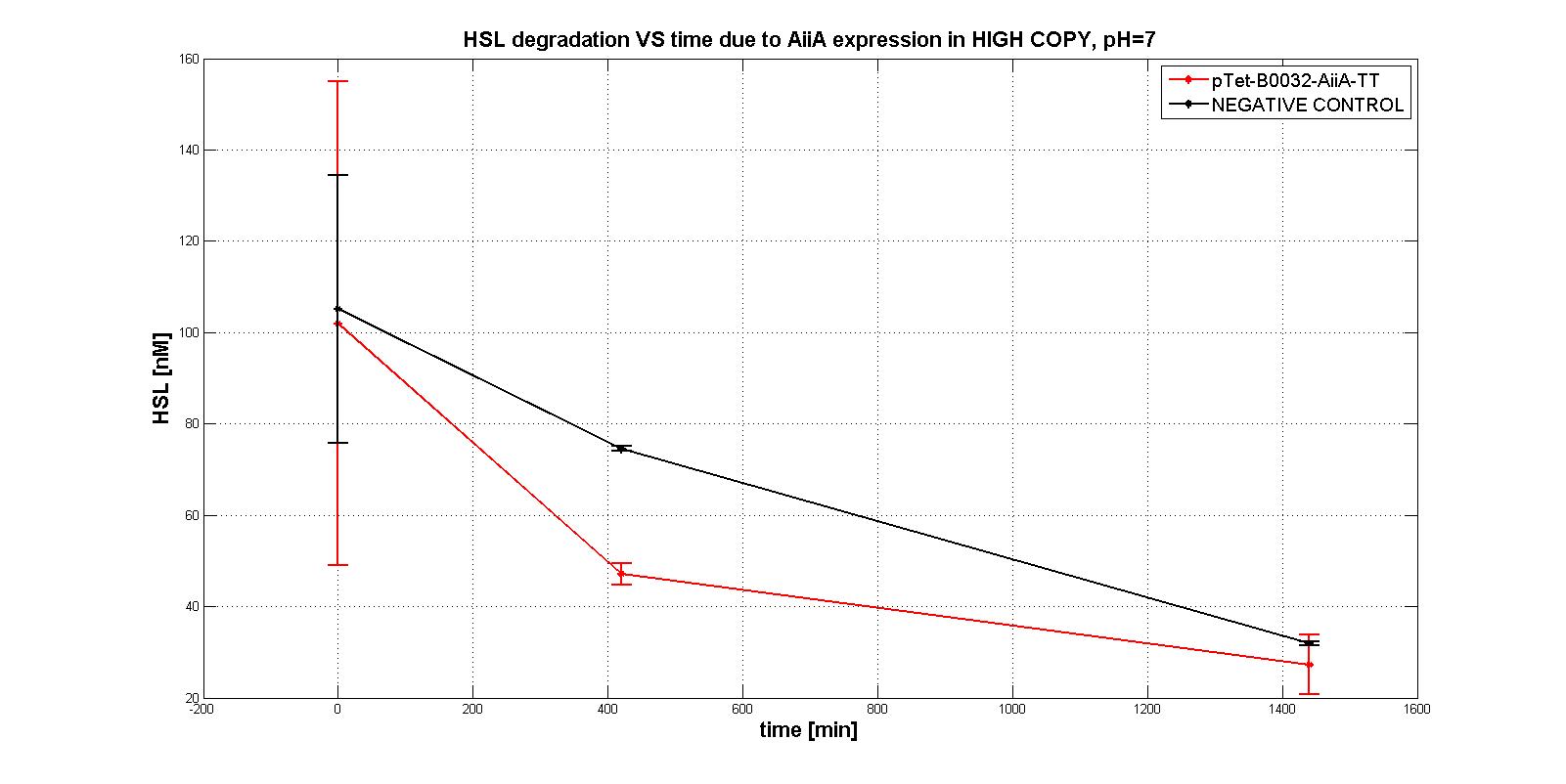
| − | One of the fundamental Biobrick® of Erwinions is the aiiA gene, it codifies for an enzyme that catalyzes the degradation of N-Acyl homoserine lactones, which are quorum sensing autoinducers. The part was published on iGEM's Parts Registry in 2013, but up until now, it's not known whether this gene can be expressed in <i>Erwinia amylovora</i> or not. Two types of aiiA were used in this project: the one published in iGEM's catalog, and the one that we synthesized thanks to sponsorship granted by IDT®. The sequence of the first mentioned is reported as being putative, while the latter's confirmed to codify for our protein of interest; nonetheless, no information is known about the expression of either in <i>Erwinia amylovora</i>. The contribution of this characterization to the existing part (BBa_C0060) as a bronze requisite is supported by the work done with it within the project. In the end, it was concluded that our composite BioBrick® (BBa_K2471000) that was created in order to express the Registry's aiiA, successfully works in <i>Erwinia amylovora</i>.
| |
| − |
| |
| − | The characterization of BBa_C0060 was carried out, first, by creating a composite part (BBa_K2471000) capable of expressing the biopart of interest; this was done by the addition of a T7 promoter and RBS (BBa_K525998) and a T1 terminator (BBa_B0010) to the basic part. The design was first done in silico utilizing SnapGene®, where the program indicated that the plasmid would have a length of 3,010 base pairs. After verifying the feasibility of creating this BioBrick®, the part was synthesized using the 3A assembly method. Agarose gel (1%) electrophoresis was performed to corroborate that the obtained molecular weight coincided with the expected one.
| |
| − |
| |
| − | <center><"https://static.igem.org/mediawiki/2017/4/45/T--Tec-Chihuahua--Snap_1.png"></center>
| |
| − | <center><div class="row">
| |
| − | <div class="col-md-8">
| |
| − | <sub><b>Figure 1. </b>(A) SnapGene® map of BBa_K2471000. (B) Agarose gel (1%) electrophoresis of BBa_K2471000 compared with NEB Quick-Load® Purple 2-Log DNA Ladder (0.1 - 10.0 kb), where the highlighted bands correspond to approximately 3,000 base pairs.</sub>
| |
| − | </div><br><br>
| |
| − | </div></div>
| |
| − |
| |
| − | The bacteria <i>Erwinia amylovora</i> was transformed by electroporation, with this new BioBrick®. Electrophoresis in polyacrylamide gel (12%) was performed in order to corroborate that the protein of interest was indeed expressed, as this is what was theoretically proposed based on the previous biographical review. The general methodology (to see specifications visit <a href="http://2017.igem.org/Team:Tec-Chihuahua/protocols">protocols section</a>) involved a growth curve of the bacteria; also, a sample was taken every hour (0 - 8 hours) and stored in refrigeration to later perform total protein extraction; the protein molecular weight marker used was BIO-RAD Precision Plus Protein™ Dual Color Standards.</p>
| |
| − |
| |
| − | <p align="justify">to calculate the molecular weight of the protein, the sequence provided by the iGEM Parts Registry for BBa_C0060 was taken and introduced in ExPASy - Translate tool, which generated the open reading frame in amino acids. This peptidic sequence was then analyzed in ExPASy - ProtParam tool, where a molecular weight of 30.32026 kDa was calculated.</p><br>
| |
| − | <center><img src="https://static.igem.org/mediawiki/2017/1/1d/T--Tec-Chihuahua--Contribution_1.png" width="700"></center>
| |
| − | <p align="center"><sub><b>Figure 2.</b>SDS-PAGE (12%) of <i>Erwinia amylovora</i> transformed with BBa_K2471000.</sub></p><br>
| |
| − | <p align="justify">As it can be seen in figure 1, there is an appreciable band present at the approximate weight of 30 kDa from the fourth hour onwards. As a point of comparison, SDS-PAGE of wild <i>Erwinia amylovora</i> was also carried out. However, no banding was seen whatsoever. We think that this happened because the protocol used is not specifically designed for this bacteria, and since <i>Erwinia amylovora</i> forms a very complex biofilm (Koczan <i>et al.</i>, 2009), the cell lysis was not carried out correctly; and thus, neither did the protein extraction. In like manner, we propose that the SDS-PAGE shown in figure 1, where the protein of interest is expressed, was done correctly because of the protein's function, which is quorum quenching; this affects the production of important components of its biofilm, which consequently leads to its deficient production. When reviewing the literature on this, not much was found, <i>Erwinia amylovora</i> is a poorly studied bacteria.
| |
| − | </p><br>
| |
| − | <palign="justify">Nonetheless, a comparison was made with an SDS-PAGE that was executed using the same methodology, but utilizing <i>Erwinia amylovora</i> transformed with BBa_K2471003 instead. As BBa_C0060 is registered as being a putative N-Acyl homoserine lactonase aiiA, this new BioBrick contains the genetic circuitry to correctly express our protein, whose sequence (that has small changes compared to BBa_C0060, with a molecular weight of 29.45340 kDa) is confirmed to indeed codify for a N-Acyl homoserine lactonase aiiA. This part is a new addition to the Parts Registry, more information about it can be found in the silver section of our results.</p><br>
| |
| − | <center><img src="https://static.igem.org/mediawiki/2017/f/f0/T--Tec-Chihuahua--Contribution_2.png" width="700"></center>
| |
| − | <p align="center"><sub><b>Figure 3.</b>SDS-PAGE (12%) of <i>Erwinia amylovora</i> transformed with BBa_K2471003.</sub></p><br>
| |
| − | <p align="justify">In the same way as figure 1, figure 2 shows an appreciable band present at the approximate weight of 30 kDa, starting from the fourth hour onwards; meaning that this band is indeed the enzyme aiiA. With this, we establish that we characterized BBa_C0060 by proving that <i>Erwinia amylovora</i> can indeed express it.</p><br><br>
| |
| − | <div class="container" style="background-color:#ccd9ff;">
| |
| − | <b> References</b><br><br>
| |
| − |
| |
| − | <p align="justify"><b>Koczan JM, McGrath MJ, Zhao Y, Sundin GW </b>(2009) Contribution of <i>Erwinia amylovora</i> exopolysaccharides amylovoran and levan to biofilm formation: implications in pathogenicity. Phytopathology 99:1237-1244 </p></div><br><br>
| |
| − | </div>
| |
| | | | |
| | | | |
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.