Difference between revisions of "Part:BBa I714891"
| (36 intermediate revisions by 2 users not shown) | |||
| Line 8: | Line 8: | ||
</gallery> | </gallery> | ||
| + | [http://2018.igem.org/Team:AFCM-Egypt# Team:AFCM-Egypt 2018] provided specific characterization for this part by using eGFP for experimental characterization of lentiviral transfer plasmid transfection efficacy and apoptotic effect on colorectal cancer cell line RKO. | ||
| + | https://static.igem.org/mediawiki/parts/f/ff/GFPAFCM18.png | ||
| + | |||
| + | |||
| + | [https://parts.igem.org/Part:BBa_K2559005# Team:SCAU-China 2018] | ||
| + | <p>The [https://parts.igem.org/Part:BBa_K2559005# BBa_K2559005] is a amended eGFP coding part improved from BBa_l714891. | ||
| + | ===Usage and Biology=== | ||
| + | The part BBa_K2559005 has a sequence improvement on the basic part submitted by iGEM07_Peking (BBa_I714891) which encodes the SDY_eGFP. However, we found out a 16 bp nucleotides redundancy in the eGFP starting coding region in BBa_I714891, after checking the sequence of BBa_I714891 from NCBI. Therefore, we decided to delete the redundant 16 bp nucleotides in BBa_I714891 to amend the length of eGFP coding sequence. The amended eGFP coding biobrick is the BBa_K2559005. | ||
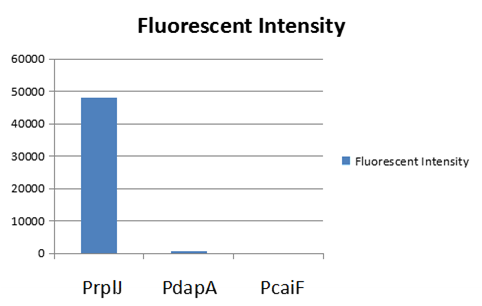
| + | To test the function of BBa_K2559005, we designed a new E.coli expression vector containing our new part termed as [https://parts.igem.org/Part:BBa_K2559003# BBa_K2559003]under a strong E.coli endogenous promoter (PrplJ). Therefore, the amended eGFP in BBa_K2559005 was driven by PrplJ promoter, and expressed in DH10B. In addition, we also applied the BBa_K2559005 in the promoter intensity analysis of our other two new parts, the [https://parts.igem.org/Part:BBa_K2559004# BBa_K2559004] and [https://parts.igem.org/Part:BBa_K2559011# BBa_K2559011] which are relatively weaker E.coli endogenous promoters (PdapA and PcaiF) (Figure 1). | ||
| + | [[File:Scau-china-2018-11.png|800px|thumb|center]] | ||
| + | |||
| + | |||
| + | [[File:Scau-china-2018-12.png|800px|thumb|center|Figure 1: Fluorescent intensity of amended eGFP driven by by PrplJ, PdapA, PcaiF promoter. | ||
| + | ]] | ||
| + | |||
| + | |||
| + | |||
| + | We summarized that our improvedpart, the amended eGFP coding biobrick BBa_K2559005 worked well in DH10B. We also hoped that our improvement on the BBa_I714891 can help their future applications by other groups in the future. However, it is difficult for us to perform additional experiments with BBa_K2559005 and BBa_I714891 due to the unavailable BBa_I714891. | ||
| + | |||
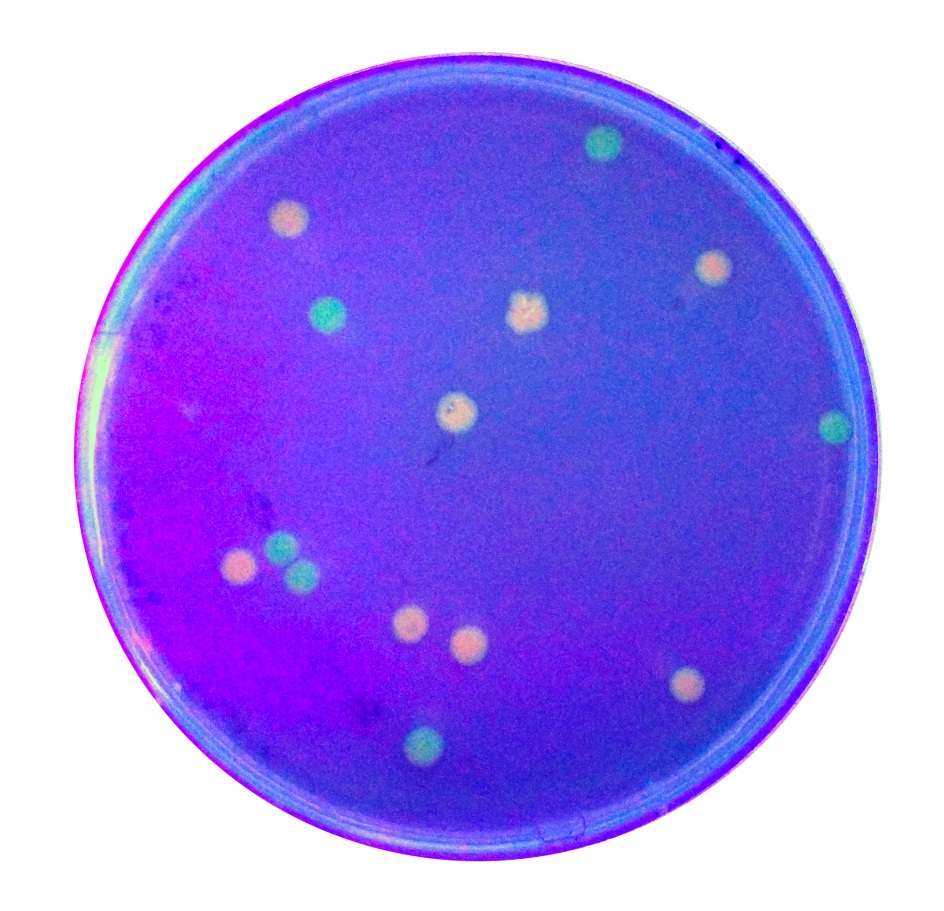
| + | To expand the application of BBa_K2559005, we searched the[https://parts.igem.org/Part:BBa_J04450# BBa_J04450]stored in registry and do another improvement in the BBa_J04450. The BBa_J04450 is a strong RFP expression vector in E.coli. As the main page of BBa_J04450 mentioned, the E.coli colonies with BBa_J04450 were in red color under normal light after about 18 hour culture on LB plate (Figure 2). We used the BBa_K2559005 to replace the RFP region in BBa_J04450, the modified part is [https://parts.igem.org/Part:BBa_K2559009# BBa_K2559009]. | ||
| + | We transferred the BBa_K2559009 to DH5α by heat-shock, and found that the fluorescence signal can be observed under the UV (Figure 2). | ||
| + | |||
| + | [[File:T-SCAU-China-Red_and_green_colonies.jpg|200px|thumb|center|Figure 2 : Figure 2 Colonies with BBa_J04450 (red colony) and BBa_K2559009 (green colony) were visualized under UV lightbox | ||
| + | ]] | ||
| + | |||
| + | So, we confirm that our improved part BBa_K2559005 can work in different E.coli expression system. We are also looking forward to more application of the BBa_K2559009! | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
| + | |||
===Usage and Biology=== | ===Usage and Biology=== | ||
Latest revision as of 13:09, 16 October 2018
SDY_EGFP
a strong reporter
[http://2018.igem.org/Team:AFCM-Egypt# Team:AFCM-Egypt 2018] provided specific characterization for this part by using eGFP for experimental characterization of lentiviral transfer plasmid transfection efficacy and apoptotic effect on colorectal cancer cell line RKO.

The BBa_K2559005 is a amended eGFP coding part improved from BBa_l714891.
Usage and Biology
The part BBa_K2559005 has a sequence improvement on the basic part submitted by iGEM07_Peking (BBa_I714891) which encodes the SDY_eGFP. However, we found out a 16 bp nucleotides redundancy in the eGFP starting coding region in BBa_I714891, after checking the sequence of BBa_I714891 from NCBI. Therefore, we decided to delete the redundant 16 bp nucleotides in BBa_I714891 to amend the length of eGFP coding sequence. The amended eGFP coding biobrick is the BBa_K2559005. To test the function of BBa_K2559005, we designed a new E.coli expression vector containing our new part termed as BBa_K2559003under a strong E.coli endogenous promoter (PrplJ). Therefore, the amended eGFP in BBa_K2559005 was driven by PrplJ promoter, and expressed in DH10B. In addition, we also applied the BBa_K2559005 in the promoter intensity analysis of our other two new parts, the BBa_K2559004 and BBa_K2559011 which are relatively weaker E.coli endogenous promoters (PdapA and PcaiF) (Figure 1).
We summarized that our improvedpart, the amended eGFP coding biobrick BBa_K2559005 worked well in DH10B. We also hoped that our improvement on the BBa_I714891 can help their future applications by other groups in the future. However, it is difficult for us to perform additional experiments with BBa_K2559005 and BBa_I714891 due to the unavailable BBa_I714891.
To expand the application of BBa_K2559005, we searched theBBa_J04450stored in registry and do another improvement in the BBa_J04450. The BBa_J04450 is a strong RFP expression vector in E.coli. As the main page of BBa_J04450 mentioned, the E.coli colonies with BBa_J04450 were in red color under normal light after about 18 hour culture on LB plate (Figure 2). We used the BBa_K2559005 to replace the RFP region in BBa_J04450, the modified part is BBa_K2559009. We transferred the BBa_K2559009 to DH5α by heat-shock, and found that the fluorescence signal can be observed under the UV (Figure 2).
So, we confirm that our improved part BBa_K2559005 can work in different E.coli expression system. We are also looking forward to more application of the BBa_K2559009! Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]