Difference between revisions of "Part:BBa K2239005"
| (2 intermediate revisions by the same user not shown) | |||
| Line 17: | Line 17: | ||
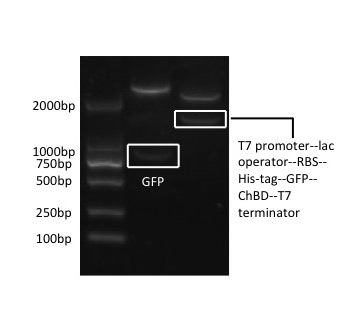
Firstly, the GFP is inserted into the modified pET-28x at BamH I and Hind III, and ChBD at Hind III and Xhol I, after proliferation in T3 vector. Then the whole gene fragment, T7 promoter--lac operator--RBS--His-tag--GFP--ChBD--T7 terminator, is retrieved from this plasmid by PCR amplification, with prefix containing EcoR I, Not I and Xba I added on its upstream primer, and suffix containing Pst I, Not I and Spe I added on its downstream primer. The PCR product is then connected to pSB1C3 at EcoR I and Pst I. | Firstly, the GFP is inserted into the modified pET-28x at BamH I and Hind III, and ChBD at Hind III and Xhol I, after proliferation in T3 vector. Then the whole gene fragment, T7 promoter--lac operator--RBS--His-tag--GFP--ChBD--T7 terminator, is retrieved from this plasmid by PCR amplification, with prefix containing EcoR I, Not I and Xba I added on its upstream primer, and suffix containing Pst I, Not I and Spe I added on its downstream primer. The PCR product is then connected to pSB1C3 at EcoR I and Pst I. | ||
| + | |||
| + | [[file:psb1c3-chbd-gfp.jpeg|300px]] | ||
[Fig. 1. pSB1C3-ChBD-GFP] | [Fig. 1. pSB1C3-ChBD-GFP] | ||
| Line 34: | Line 36: | ||
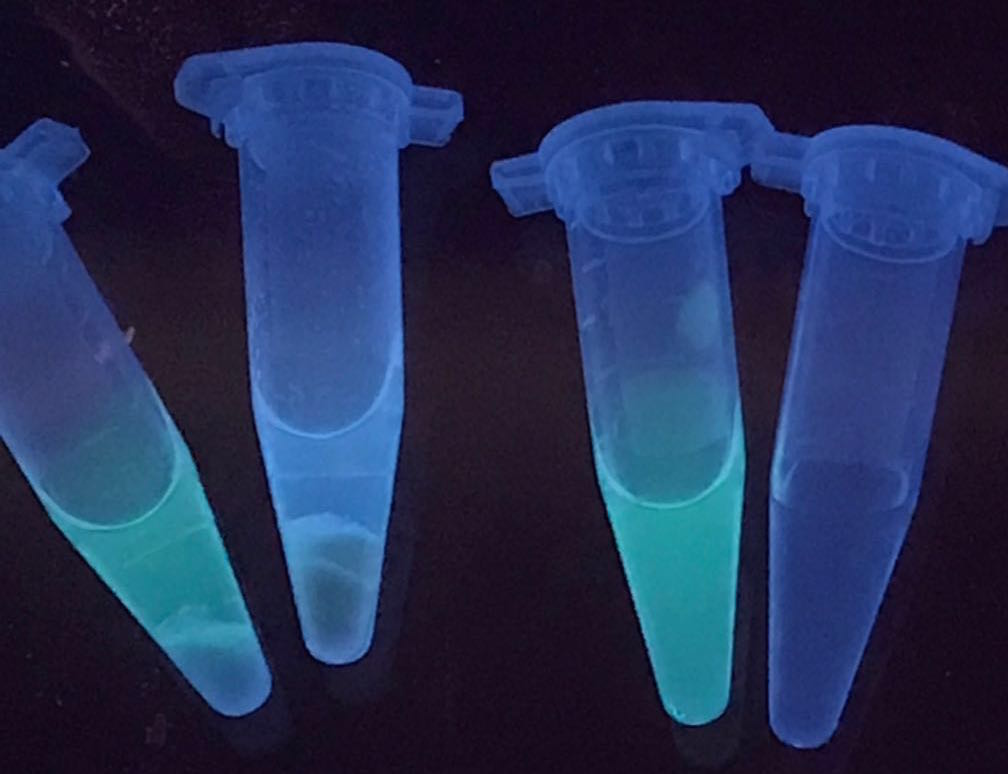
Tube 3 contains GFP-ChBD, with a significant green color in the solution. After adding chitin powder inside the tube, the color in the solution disappears and shows on the chitin powder, as presented in tube 2. It proves that the ChBD successfully binds the GFP onto the chitin powder. In contrast, the color of the solution is not changed when adding chitin powder into the GFP solution without ChBD. As a result, the ChBD is well functioned. | Tube 3 contains GFP-ChBD, with a significant green color in the solution. After adding chitin powder inside the tube, the color in the solution disappears and shows on the chitin powder, as presented in tube 2. It proves that the ChBD successfully binds the GFP onto the chitin powder. In contrast, the color of the solution is not changed when adding chitin powder into the GFP solution without ChBD. As a result, the ChBD is well functioned. | ||
| + | |||
| + | [[file:chbd.jpeg|250px]] | ||
[Fig.2. ChBD function: tube1, GFP and chitin; tube2, GFP-ChBD and chitin; tube3, GFP-ChBD; tube4, water] | [Fig.2. ChBD function: tube1, GFP and chitin; tube2, GFP-ChBD and chitin; tube3, GFP-ChBD; tube4, water] | ||
| Line 40: | Line 44: | ||
<h3>Reference</h3> | <h3>Reference</h3> | ||
[1] Takahisa Ikegami, Terumasa Okada, Masayuki Hashimoto, Shizuka Seino, Takeshi Watanabe, and Masahiro Shirakawa: Solution Structure of the Chitin-binding Domain of Bacillus circulans WL-12 Chitinase A1. The Journal Of Biological Chemistry, 2000. | [1] Takahisa Ikegami, Terumasa Okada, Masayuki Hashimoto, Shizuka Seino, Takeshi Watanabe, and Masahiro Shirakawa: Solution Structure of the Chitin-binding Domain of Bacillus circulans WL-12 Chitinase A1. The Journal Of Biological Chemistry, 2000. | ||
| + | |||
<!-- --> | <!-- --> | ||
Latest revision as of 09:55, 1 November 2017
ChBD-GFP
ChBD-GFP (T7 promoter--lac operator--RBS--His-tag--GFP--ChBD--T7 terminator)
This device codes for the GFP-ChBD fusion protein.
Construct
The vector of GFP-ChBD for its expression is pET-28x. It is formed by modifying the restriction enzyme sites EcoR I and Xba I of vector pET-28a.
The GFP sequence is retrieved from the GenBank. It is artificially synthesized and inserted into plasmid pUC57. The GFP gene is then cloned from the plasmid by PCR amplification. The restriction site BamH I is added to the upstream primer, and Hind III is added to the downstream primer.
The ChBD sequence is retrieved from the GenBank. It is artificially synthesized and inserted into plasmid pUC57. The ChBD gene is then cloned from the plasmid by PCR amplification, with the restriction site Hind III added to the upstream primer, and Xhol I added to the downstream primer.
Firstly, the GFP is inserted into the modified pET-28x at BamH I and Hind III, and ChBD at Hind III and Xhol I, after proliferation in T3 vector. Then the whole gene fragment, T7 promoter--lac operator--RBS--His-tag--GFP--ChBD--T7 terminator, is retrieved from this plasmid by PCR amplification, with prefix containing EcoR I, Not I and Xba I added on its upstream primer, and suffix containing Pst I, Not I and Spe I added on its downstream primer. The PCR product is then connected to pSB1C3 at EcoR I and Pst I.
[Fig. 1. pSB1C3-ChBD-GFP]
Usage and Biology
This part is used to test the function of ChBD (chitin binding domain), which is able to bind to chitin.[1]
Expression
The constructed pET28x-GFP-ChBD plasmid is transformed into BL21(DE3) E.coli for expression. After that, when the OD 600 reached 0.6-0.8, 0.2mM IPTG is added in the liquid culture. The mixture is shaken at 20 ℃ overnight. The bacteria is collected by centrifugation at low temperature, 8000 rpm for 10 minutes, and the supernatant is discarded. The bacteria is then resuspended using 0.15M pH8.8 Tris-HCL, and is broken by ultrasonication.
Proof of the ChBD function
The resulted solution after expression is mixed with chitin powder. Also, the solution with GFP-ChBD but without chitin, solution with the presence of GFP and chitin, and water in comparison, are presented.
Tube 3 contains GFP-ChBD, with a significant green color in the solution. After adding chitin powder inside the tube, the color in the solution disappears and shows on the chitin powder, as presented in tube 2. It proves that the ChBD successfully binds the GFP onto the chitin powder. In contrast, the color of the solution is not changed when adding chitin powder into the GFP solution without ChBD. As a result, the ChBD is well functioned.
[Fig.2. ChBD function: tube1, GFP and chitin; tube2, GFP-ChBD and chitin; tube3, GFP-ChBD; tube4, water]
Reference
[1] Takahisa Ikegami, Terumasa Okada, Masayuki Hashimoto, Shizuka Seino, Takeshi Watanabe, and Masahiro Shirakawa: Solution Structure of the Chitin-binding Domain of Bacillus circulans WL-12 Chitinase A1. The Journal Of Biological Chemistry, 2000.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 999
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 751
Illegal BsaI.rc site found at 833