Difference between revisions of "Part:BBa K2232014"
James zhang (Talk | contribs) |
|||
| (2 intermediate revisions by 2 users not shown) | |||
| Line 17: | Line 17: | ||
<partinfo>BBa_K2232014 parameters</partinfo> | <partinfo>BBa_K2232014 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | ===iGEM2017 SZU-China=== | ||
| + | To realize the self-healing of cracks in concrete, we need to increase the mineralization capacity of B.subtilis. The Healer in our project is Carbonic anhydrase(CA) , which catalyzes the hydration of CO<sub>2</sub> to produce HCO<sub>3</sub><sup>-</sup> and captures free Ca<sup>2+</sup> with OH<sup>-</sup> in the environment to form Calcium carbonate precipitation. | ||
| + | The new part TSLV1-CA (BBa_K2232014) expresses and functiones intracellularly. We constructed a shuttle vector to transform this part and the positive clones was confirmed by nucleic acid electrophoresis(Fig.1). | ||
| + | <div> | ||
| + | <center><html><img src='https://static.igem.org/mediawiki/parts/d/d0/TSLV1-CA1.jpeg' style="width:30%;margin:0 auto"> | ||
| + | <center>Fig.1 1% Agarose Gel Electrophoresis of Vector_ TSLV1-CA and its identification by restriction digestion. Lane 1: Complete plasmid; Lane 2: Plasmid digested by KpnI and HindIII; Lane M: DL marker.The length of part TSLV1-CA was 949 bp and the blank vector was 6785 bp.</center> | ||
| + | </html></center> | ||
| + | </div> | ||
| + | |||
| + | The crude enzyme solution was obtained by cell disruption using ultrasonic, followed by SDS-PAGE protein electrophoresis and Coomassie blue staining(Fig.2). | ||
| + | <div> | ||
| + | <center><html><img src='https://static.igem.org/mediawiki/parts/5/50/TSLV1-CA2.png' style="width:30%;margin:0 auto"> | ||
| + | <center>Fig.2 SDS-PAGE analysis of endocellular protein of original B.subtilis and the transformant of CA. | ||
| + | Lane M: Marker ladder; Lane 1: Modified strain WB800_ TSLV1-CA; Lane 2: Modified strain WB800_ OF4-CA; Lane 3: Original strain WB800. Lane 1 and lane 2 have a band of 35~37kd respectively (in red box), which correspond with molecular weight of TSLV1-CA (35kDa) and OF4-CA (34.5kDa). | ||
| + | |||
| + | </center> | ||
| + | </html></center> | ||
| + | </div> | ||
| + | |||
| + | For determining the activity of CA, hydration of CO<sub>2</sub> was measured using electrometric Wilbur–Anderson assay according to Khalifah et al. (1991) with certain modifications. | ||
| + | The assay was performed at 4 °C by adding 0.5 mL of the crude enzyme solution (0.5 ml distilled water in blank group) to 10 mL of 30mM PBS (pH 8.0). The reaction was initiated by adding 5.0 mL of ice-cold CO<sub>2</sub> saturated water. The time interval for the pH to drop by 1.5 unit (from 8.0 to 6.5) due to protons released during hydration of CO2 was measured. The reactions were performed in triplicates and average of three replicates was used in calculations. | ||
| + | We calculated the activity according to the formula U= (T0 –T1)/ T0, where T0 and T1 represent time for pH change of blank group and samples group respectively. The CA activity was shown in Fig.3. | ||
| + | <div> | ||
| + | <center><html><img src='https://static.igem.org/mediawiki/parts/2/2c/TSLV1-CA4.png' style="width:50%;margin:0 auto"> | ||
| + | <center>Fig.3 CA activity of crude enzyme solution from measured by Brownell’s method | ||
| + | </center> | ||
| + | </html></center> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | ===iGEM2022 UCL=== | ||
| + | <h1>Description</h1> | ||
| + | This was the original carbonic anhydrase part from SZU-China 2017 iGEM team (Part: <partinfo>BBa_K2232014</partinfo>). We have mutated the SapI site in the coding sequence and successfully cloned into our pCT5c Type IIS compatible plasmid (<partinfo>BBa_K4417000</partinfo>). | ||
| + | |||
| + | |||
| + | <h1>Usage and Biology</h1> | ||
| + | *This part can be transformed in both ''B. subtilis'' and ''E. coli''. | ||
| + | *Inducer: p-isopropyl benzoate (cumate). | ||
| + | *Cumate is non-toxic to the host. In our experiment, we induced the urease expression with 50 μM cumate. | ||
| + | *The copy number of this plasmid in ''B. subtilis'' and ''E. coli'' is unknown. | ||
| + | *NdeI restriction can be used to change the promoter. | ||
| + | |||
| + | <h1>Cloning Strategy</h1> | ||
| + | This part was cloned into pCT5c (<partinfo>BBa_K4417000</partinfo>) using restriction digest. | ||
| + | |||
| + | [[File:CA17 in pCT5c.jpg|600px|thumb|center|'''Figure 2:''' Construction of P43-RBS-CA17-rrnB T1 Terminator in pCT5c.]] | ||
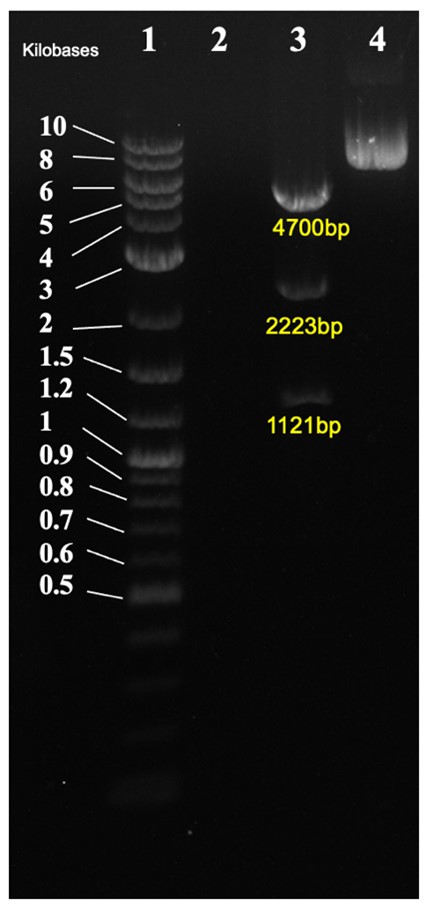
| + | In Figure 3, the. Cloned plasmid was checked by diagnostic digest. Correct band size was observed of 4700bp, 2223bp, and 1121bp. | ||
| + | |||
| + | [[File:CA17 gel.jpg|600px|thumb|center|'''Figure 3:''' Diagnostic digest of CA17. 1: DNA ladder, 2: empty, 3: CA22 cut with KpnI/PstI/SpeI, 4: CA22 uncut.]] | ||
| + | |||
| + | <h1>Characterization</h1> | ||
| + | |||
| + | '''SDS Page''' | ||
| + | |||
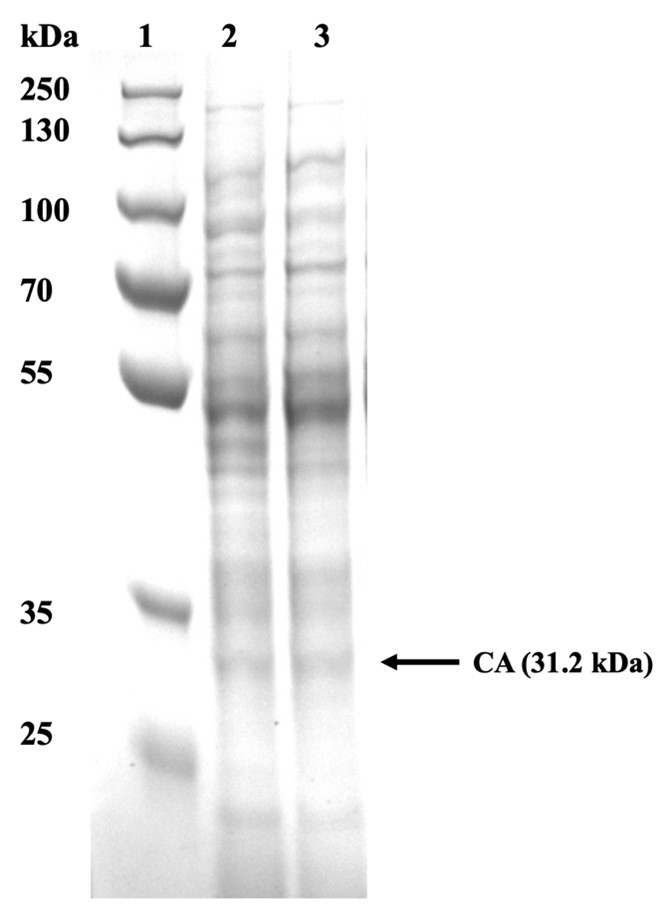
| + | In order to observe whether the CA22 was successfully expressed, we analysed our cell pellet using SDS PAGE. The pellet obtained from the 10 mL cultures was then resuspended in Tris Buffer Saline at an OD600. Once resuspended, the sample was cell lysed using sonication. Following sonication, the samples were spanned to separate the soluble and insoluble fragments from the whole cell lysate. 60 μL from each sample were obtained and stained with Laemmli reagent. | ||
| + | [[File:CA17 protein gel.jpg|600px|thumb|center|'''Figure 4:''' SDS PAGE of CA22 in ''E. coli''. All the strains were grown in LB medium; 1: PageRuler Protein Ladder, 2: WT ''E. coli'' cell lysate, 3: CA22 cell lysate.]] | ||
| + | From Figure 4, it could be concluded that the urease was successfully produced since the correct bands are observed in cell lysate samples. Carbonic anhydrase was identified at 31.2 kDa. | ||
| + | |||
| + | '''Carbonic anhydrase assay''' | ||
| + | |||
| + | We measured activity of our bacteria engineered to overexpress carbonic anhydrase (CA). We are interested in CA because it catalyzes hydration of CO2, but it can also act as an ester hydration catalyst. Combining clarified cell lysate (of cultures with OD<sub>600</sub> = 1.74) with 4-NPA, a non-physiological commercially available ester, we were able to prove the CA activity. CA catalyzes hydration of this ester which results in yellowing of the solution. The amount of yellowing is quantified by OD<sub>348</sub> measurements corrected for OD<sub>600</sub> of cell cultures before lysing. | ||
| + | |||
| + | As the original assay was designed to be performed with purified protein and we didn’t have it, we decided to run it with different concentrations of clarified cell lysate to ensure proper assay sensitivity. Ultimately, we have shown that the CA17 genetic construct upregulates carbonic. | ||
| + | |||
| + | [[File:Carbonic anhydrase assay for WT ''E. coli'' and transformed CA17.jpg|600px|thumb|center|'''Figure 5:''' Carbonic anhydrase assay for WT E. coli and transformed CA17.]] | ||
| + | |||
| + | '''Biocementation''' | ||
| + | |||
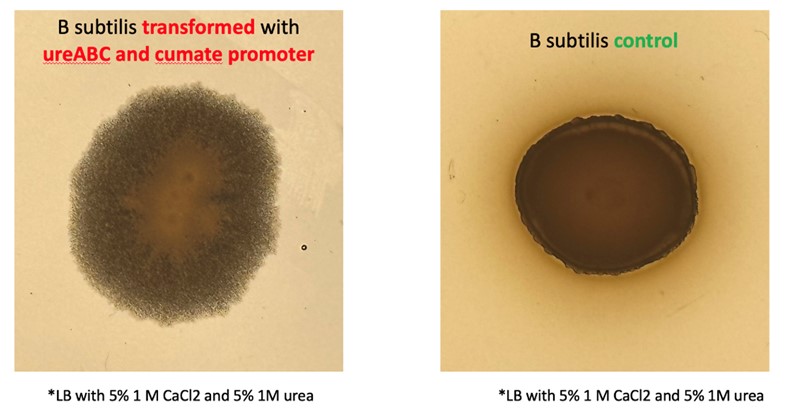
| + | Induced engineered bacteria should precipitate significantly more calcium carbonate in calcium rich environments than wild type bacteria. The first place where this can be observed is on the cell colony morphology. Our engineered bacteria produce crystal-like colonies (on calcium rich LB plates) which points to increased CaCO<sub>3</sub> precipitation caused by our genetic constructs. | ||
| + | |||
| + | [[File:Ca plate.jpg|600px|thumb|center|'''Figure 6:''' Precipitation on calcium rich agar plates by CA17.]] | ||
| + | |||
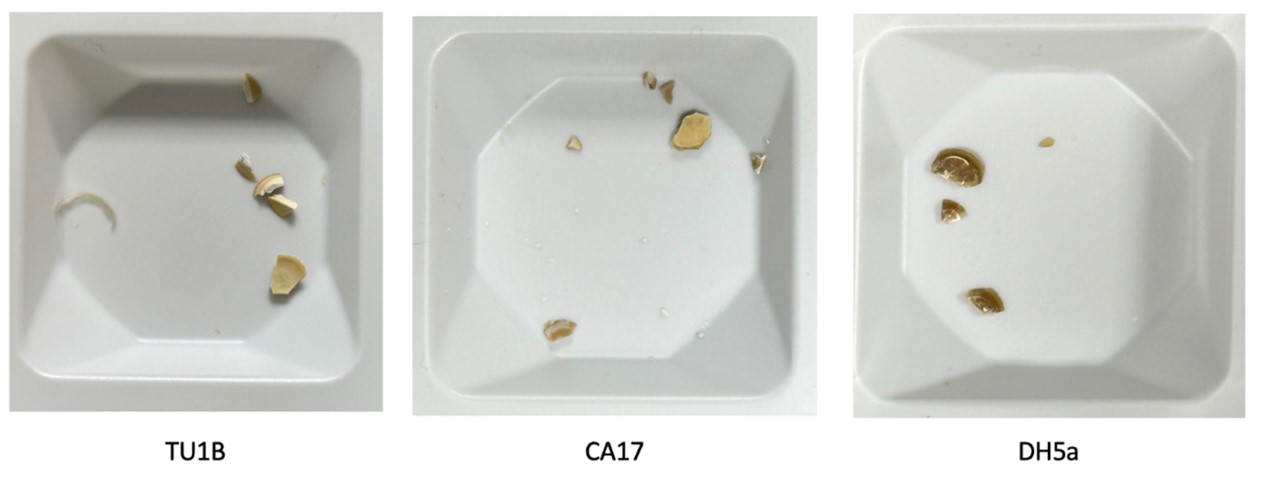
| + | The dried aggregate of transformed bacteria is pale and matte compared with the aggregate of wild type bacteria, which is dark brown and has a glass-like surface. This is an indication of enhanced calcium carbonate precipitation. | ||
| + | [[File:dry plates.jpg|600px|thumb|center|'''Figure 7:''' Dry aggregates from biocementation.]] | ||
| + | |||
| + | We measured the dry weight of aggregates formed by the engineered and wild type cells in a calcium and urea rich medium. There is a statically significant (p<0.05) increase in weight between the engineered cells (TU1B, CA17) and the wild type (DH5a). The increase in dry weight of the aggregate also points to enhanced calcium carbonate precipitation of the engineered cells. | ||
| + | [[File:dry weight.jpg|600px|thumb|center|'''Figure 8:''' Dry weight of aggregates from biocementation.]] | ||
| + | |||
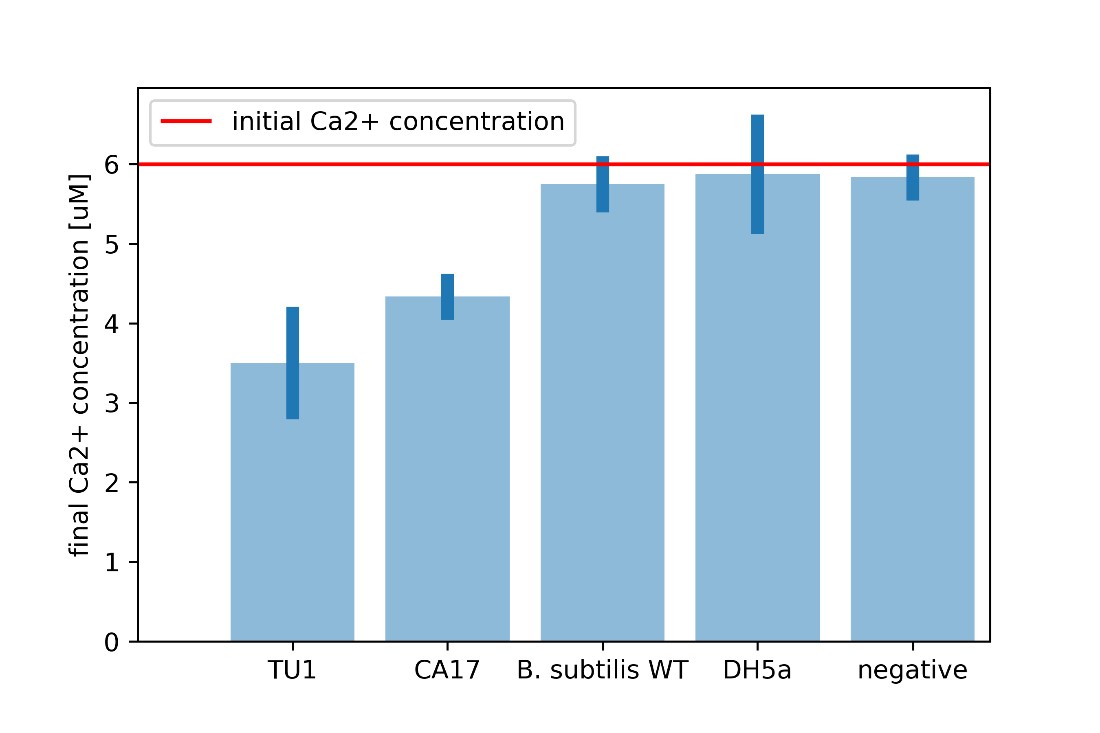
| + | For every calcium carbonate molecule precipitated, one calcium ion leaves the solution. Calcium carbonate is insoluble in water and hence, this calcium ion can no longer interact with other chemicals in the solution. If we know the initial calcium ion concentration, we can perform a Patton-Reeder calorimetric assay to find the resulting calcium concentration. The difference between the initial and final calcium concentrations equates the calcium that left the solution either to the cell or to be bound to calcium carbonate. | ||
| + | [[File:Ca conc.jpg|600px|thumb|center|'''Figure 9:''' Calcium ion assay.]] | ||
| + | |||
| + | It is clear that the CA17 uptakes significantly more calcium ions from the environment, combined with the observed increase in the dry aggregate weight, this proves enhanced CaCO<sub>3</sub> precipitation. | ||
Latest revision as of 15:27, 12 October 2022
P43_TSLV1-CA-alpha_rrnBT1
For facilitating the interconversion of CO2 and HCO3− which is beneficial to the precipitation of CaCO3, a plasmid containing Carbonic anhydrase coding gene have built and transferred into B. subtilis strain WB800 and express successfully.The promoter P43 used in the plasmid is a strong promoter in Bacillus subtilis 168,and the Carbonic anhydrase is from The polyextremophilic bacterium Bacillus halodurans TSLV1 (MTCC 10961, 16S rDNA Acc. No. HQ235051) which belong to α-type in classes of CAs. The terminator we used is rrnBT1(BBa_B0010) from E. coli rrnB, which will stop the Transcription of gene and raise the efficiency of expression.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 851
Illegal NotI site found at 1138 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 831
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 376
Illegal SapI.rc site found at 534
Illegal SapI.rc site found at 846
iGEM2017 SZU-China
To realize the self-healing of cracks in concrete, we need to increase the mineralization capacity of B.subtilis. The Healer in our project is Carbonic anhydrase(CA) , which catalyzes the hydration of CO2 to produce HCO3- and captures free Ca2+ with OH- in the environment to form Calcium carbonate precipitation. The new part TSLV1-CA (BBa_K2232014) expresses and functiones intracellularly. We constructed a shuttle vector to transform this part and the positive clones was confirmed by nucleic acid electrophoresis(Fig.1).

The crude enzyme solution was obtained by cell disruption using ultrasonic, followed by SDS-PAGE protein electrophoresis and Coomassie blue staining(Fig.2).

For determining the activity of CA, hydration of CO2 was measured using electrometric Wilbur–Anderson assay according to Khalifah et al. (1991) with certain modifications. The assay was performed at 4 °C by adding 0.5 mL of the crude enzyme solution (0.5 ml distilled water in blank group) to 10 mL of 30mM PBS (pH 8.0). The reaction was initiated by adding 5.0 mL of ice-cold CO2 saturated water. The time interval for the pH to drop by 1.5 unit (from 8.0 to 6.5) due to protons released during hydration of CO2 was measured. The reactions were performed in triplicates and average of three replicates was used in calculations. We calculated the activity according to the formula U= (T0 –T1)/ T0, where T0 and T1 represent time for pH change of blank group and samples group respectively. The CA activity was shown in Fig.3.

iGEM2022 UCL
Description
This was the original carbonic anhydrase part from SZU-China 2017 iGEM team (Part: BBa_K2232014). We have mutated the SapI site in the coding sequence and successfully cloned into our pCT5c Type IIS compatible plasmid (BBa_K4417000).
Usage and Biology
- This part can be transformed in both B. subtilis and E. coli.
- Inducer: p-isopropyl benzoate (cumate).
- Cumate is non-toxic to the host. In our experiment, we induced the urease expression with 50 μM cumate.
- The copy number of this plasmid in B. subtilis and E. coli is unknown.
- NdeI restriction can be used to change the promoter.
Cloning Strategy
This part was cloned into pCT5c (BBa_K4417000) using restriction digest.
In Figure 3, the. Cloned plasmid was checked by diagnostic digest. Correct band size was observed of 4700bp, 2223bp, and 1121bp.
Characterization
SDS Page
In order to observe whether the CA22 was successfully expressed, we analysed our cell pellet using SDS PAGE. The pellet obtained from the 10 mL cultures was then resuspended in Tris Buffer Saline at an OD600. Once resuspended, the sample was cell lysed using sonication. Following sonication, the samples were spanned to separate the soluble and insoluble fragments from the whole cell lysate. 60 μL from each sample were obtained and stained with Laemmli reagent.
From Figure 4, it could be concluded that the urease was successfully produced since the correct bands are observed in cell lysate samples. Carbonic anhydrase was identified at 31.2 kDa.
Carbonic anhydrase assay
We measured activity of our bacteria engineered to overexpress carbonic anhydrase (CA). We are interested in CA because it catalyzes hydration of CO2, but it can also act as an ester hydration catalyst. Combining clarified cell lysate (of cultures with OD600 = 1.74) with 4-NPA, a non-physiological commercially available ester, we were able to prove the CA activity. CA catalyzes hydration of this ester which results in yellowing of the solution. The amount of yellowing is quantified by OD348 measurements corrected for OD600 of cell cultures before lysing.
As the original assay was designed to be performed with purified protein and we didn’t have it, we decided to run it with different concentrations of clarified cell lysate to ensure proper assay sensitivity. Ultimately, we have shown that the CA17 genetic construct upregulates carbonic.
Biocementation
Induced engineered bacteria should precipitate significantly more calcium carbonate in calcium rich environments than wild type bacteria. The first place where this can be observed is on the cell colony morphology. Our engineered bacteria produce crystal-like colonies (on calcium rich LB plates) which points to increased CaCO3 precipitation caused by our genetic constructs.
The dried aggregate of transformed bacteria is pale and matte compared with the aggregate of wild type bacteria, which is dark brown and has a glass-like surface. This is an indication of enhanced calcium carbonate precipitation.
We measured the dry weight of aggregates formed by the engineered and wild type cells in a calcium and urea rich medium. There is a statically significant (p<0.05) increase in weight between the engineered cells (TU1B, CA17) and the wild type (DH5a). The increase in dry weight of the aggregate also points to enhanced calcium carbonate precipitation of the engineered cells.
For every calcium carbonate molecule precipitated, one calcium ion leaves the solution. Calcium carbonate is insoluble in water and hence, this calcium ion can no longer interact with other chemicals in the solution. If we know the initial calcium ion concentration, we can perform a Patton-Reeder calorimetric assay to find the resulting calcium concentration. The difference between the initial and final calcium concentrations equates the calcium that left the solution either to the cell or to be bound to calcium carbonate.
It is clear that the CA17 uptakes significantly more calcium ions from the environment, combined with the observed increase in the dry aggregate weight, this proves enhanced CaCO3 precipitation.