Difference between revisions of "Part:BBa K2243018"
| (13 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | + | ||
<partinfo>BBa_K2243018 short</partinfo> | <partinfo>BBa_K2243018 short</partinfo> | ||
To test the influence of Bxb1 attB/P to terminator L3S3P22 (abbreviation: 322) in the forward direction. | To test the influence of Bxb1 attB/P to terminator L3S3P22 (abbreviation: 322) in the forward direction. | ||
| − | <!-- | + | <!-- --> |
| − | + | <h2>Usage</h2> | |
| + | |||
| + | We constructed this part to characterize the recombination efficiency of the recombinase Bxb1 gp35 (BBa_K2243012). It consists of the terminator L3S3P22 (abbreviation: 322) in the forward direction flanked by attB and attP sites of recombinase Bxb1 gp35. Upon recombination, the orientation of the terminator changes. As a result, expression of downstream sequence is initiated, and upstream sequence is transcribed. | ||
| + | |||
| + | <!-- --> | ||
| + | <h2>Biology</h2> | ||
| + | |||
| + | The attP site of Mycobacteriophage Bxb1 is used to integrate phage DNA at the host attB site of Mycobacterium smegmatis, generating the recombinant junctions attL and attR. DNA cleavage and re-ligation occur at the central crossover region at attB and attP, which allows the sequence to be flipped, excised, or inserted between recognition sites. We obtained the terminator, attB and attP sites by oligo synthesis. | ||
| + | |||
| + | <!-- --> | ||
| + | <h2>Characterization</h2> | ||
| + | |||
| + | We first characterized the terminator strength using the following formula: | ||
| + | |||
| + | |||
| + | Ts=〖GFP〗of the random sequence/〖[GFP]〗with the terminator | ||
| + | |||
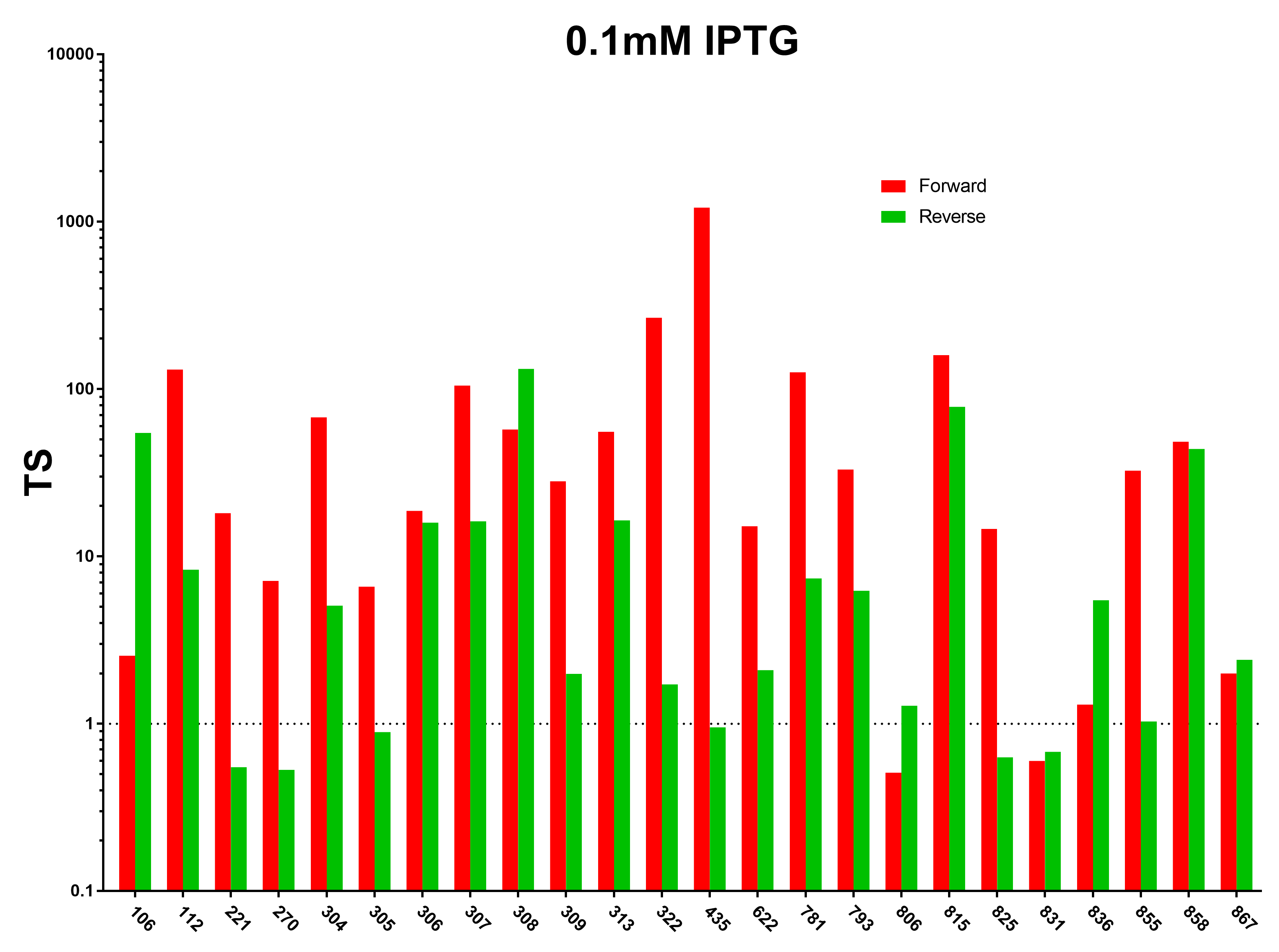
| + | [[File:Peking_TS_0.1mM.png|800px|thumb|center|Fig.1 Terminator Strength Induced with 0.1mM IPTG]] | ||
| + | |||
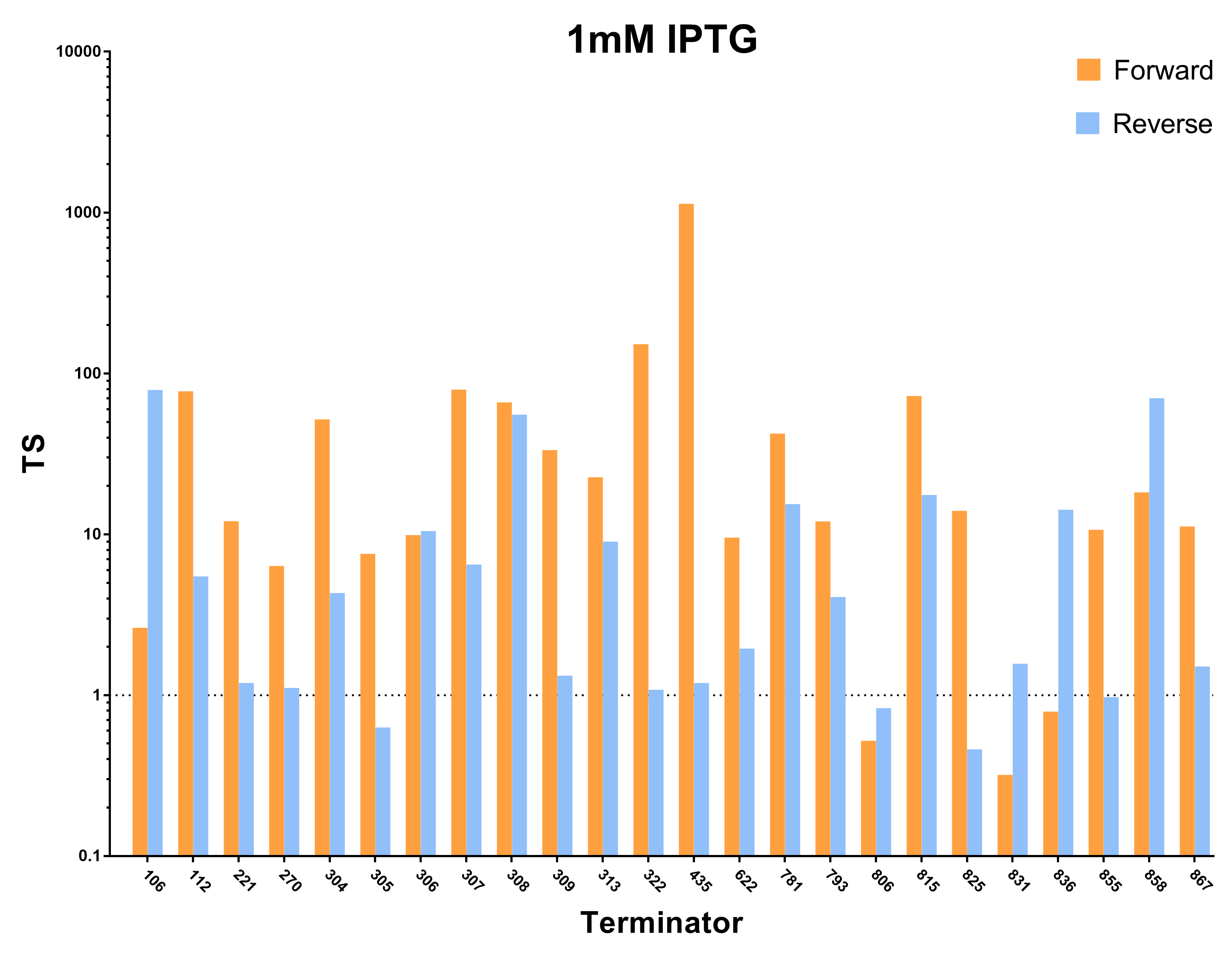
| + | [[File:Peking_TS_1mM.png|800px|thumb|center|Fig.2 Terminator Strength Induced with 1mM IPTG]] | ||
| + | |||
| + | And we sifted out 6 desirable unidirectional terminators without potential cryptic promotor in both orientations. | ||
| + | |||
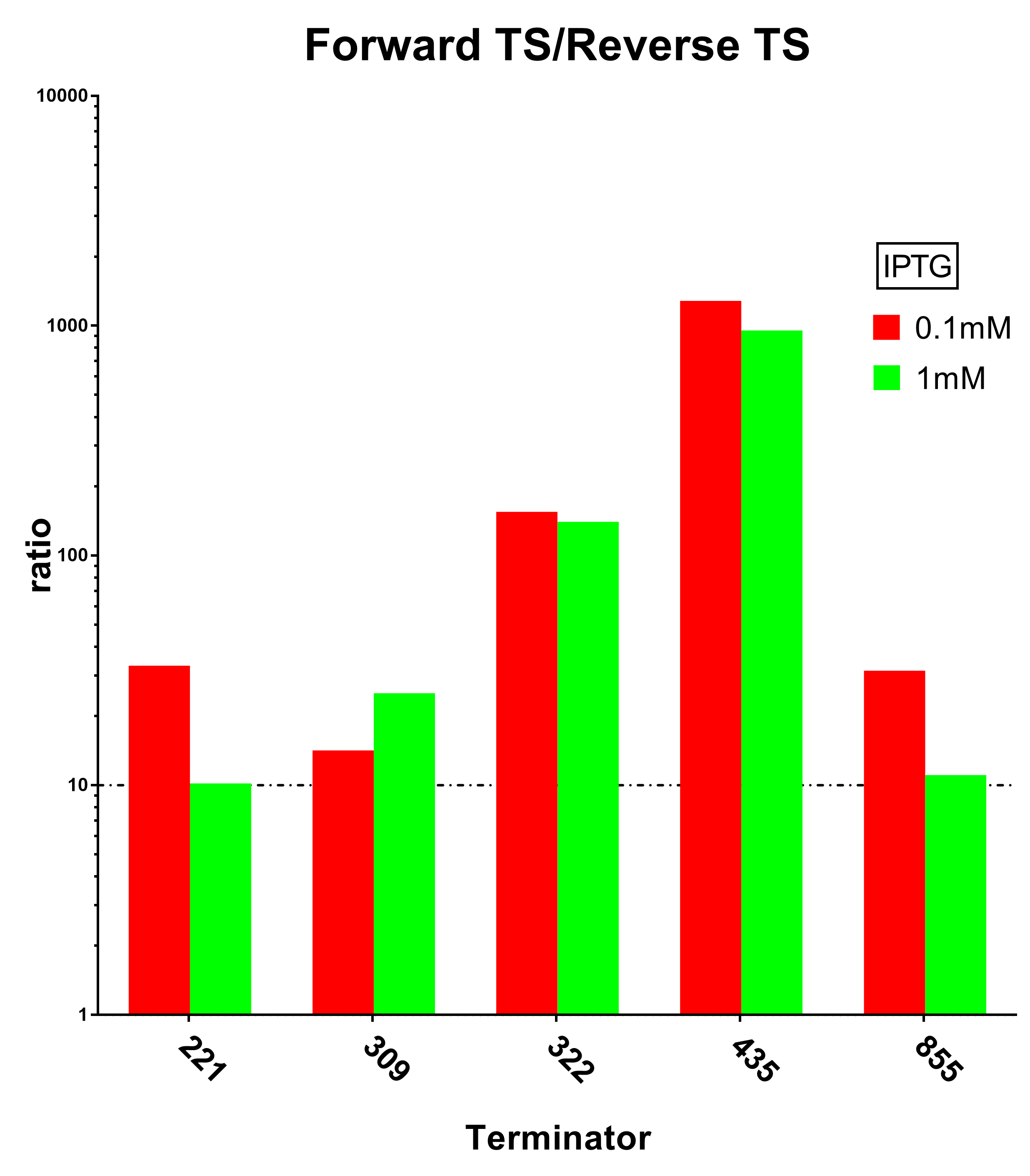
| + | [[File:Peking ratio.png|600px|thumb|center|Fig.3 Terminator Strength Induced with 0.1M IPTG]] | ||
| + | |||
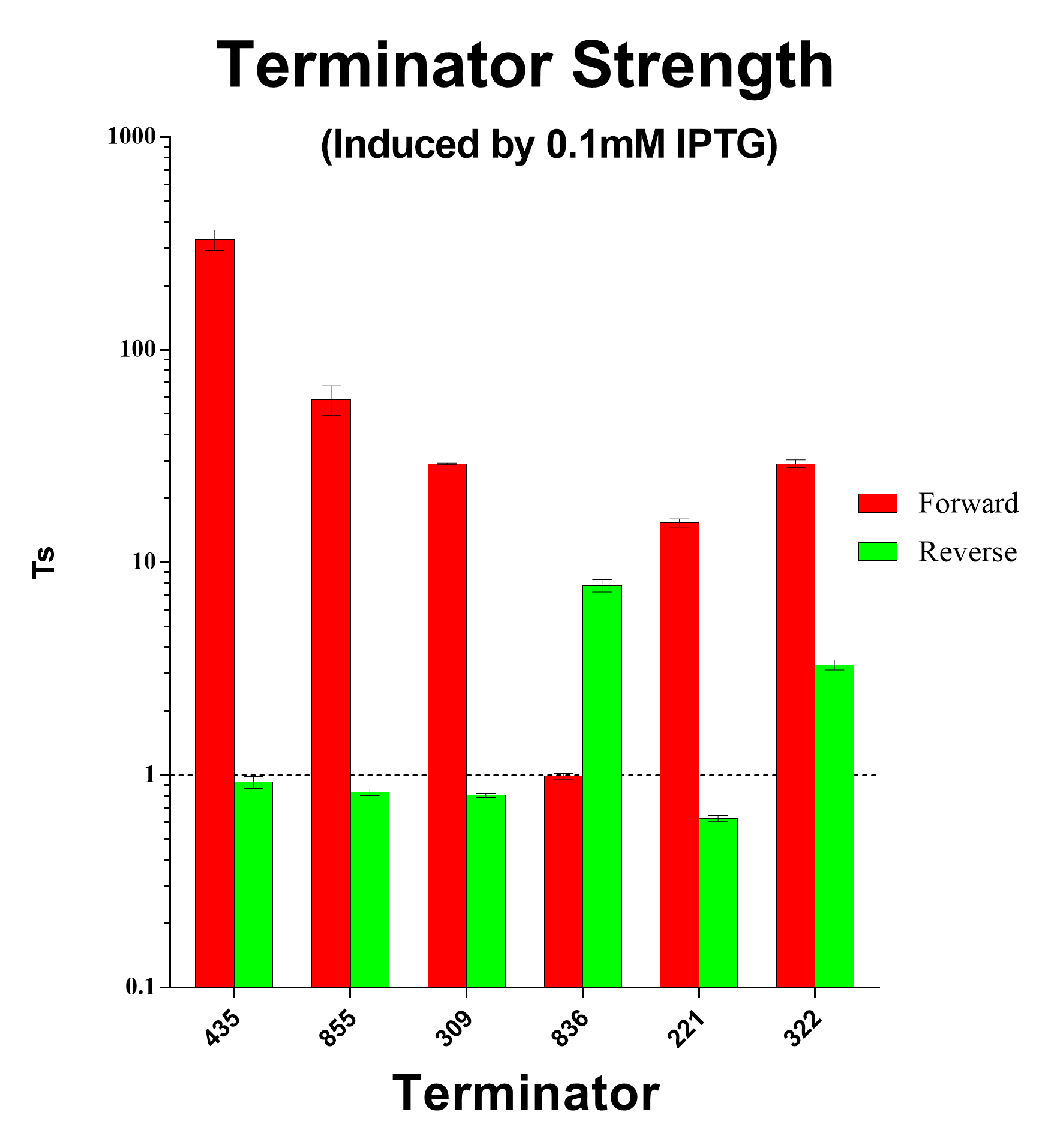
| + | [[File:Peking TS good terminators.png|600px|thumb|center|Fig.4 Forward and Reverse Terminator Strength Ratio Induced with 0.1mM and 1mM IPTG]] | ||
| + | |||
| + | <!-- --> | ||
| + | <h2>Terminator Reference Table</h2> | ||
| + | |||
| + | [[Media:Peking_trt.xlsx]] | ||
| − | |||
<!-- --> | <!-- --> | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
| + | |||
| + | |||
<partinfo>BBa_K2243018 SequenceAndFeatures</partinfo> | <partinfo>BBa_K2243018 SequenceAndFeatures</partinfo> | ||
Latest revision as of 15:43, 1 November 2017
Bxb1 attB_322F_Bxb1 attP
To test the influence of Bxb1 attB/P to terminator L3S3P22 (abbreviation: 322) in the forward direction.
Usage
We constructed this part to characterize the recombination efficiency of the recombinase Bxb1 gp35 (BBa_K2243012). It consists of the terminator L3S3P22 (abbreviation: 322) in the forward direction flanked by attB and attP sites of recombinase Bxb1 gp35. Upon recombination, the orientation of the terminator changes. As a result, expression of downstream sequence is initiated, and upstream sequence is transcribed.
Biology
The attP site of Mycobacteriophage Bxb1 is used to integrate phage DNA at the host attB site of Mycobacterium smegmatis, generating the recombinant junctions attL and attR. DNA cleavage and re-ligation occur at the central crossover region at attB and attP, which allows the sequence to be flipped, excised, or inserted between recognition sites. We obtained the terminator, attB and attP sites by oligo synthesis.
Characterization
We first characterized the terminator strength using the following formula:
Ts=〖GFP〗of the random sequence/〖[GFP]〗with the terminator
And we sifted out 6 desirable unidirectional terminators without potential cryptic promotor in both orientations.
Terminator Reference Table
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 4
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 24
Illegal BsaI site found at 103
Illegal BsaI.rc site found at 147