Difference between revisions of "Part:BBa K2285012"
(→Usage and Biology) |
|||
| (34 intermediate revisions by 2 users not shown) | |||
| Line 4: | Line 4: | ||
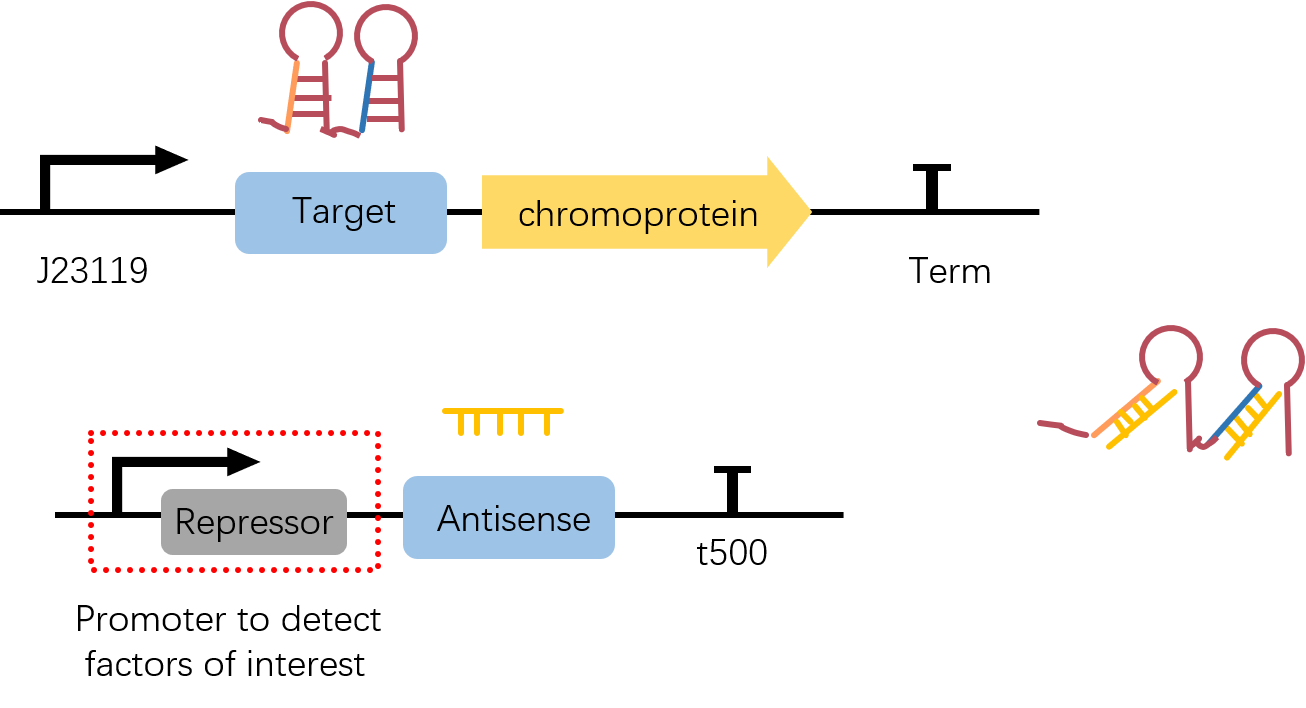
constitutive promoter J23119 followed by a stem-loop structure Target3, with chromoprotein eforRed as the reporter gene. | constitutive promoter J23119 followed by a stem-loop structure Target3, with chromoprotein eforRed as the reporter gene. | ||
| + | The original part of eforRed chromoprotein is presented on <partinfo>BBa_K592012</partinfo>. What should be noticed is, the original part is a coding part and doesn't have its own RBS and Term. Therefore, to achieve greater convenience fo our Star-chromoprotein reporter system, we add the RBS which is on the upstream of sfGFP(<partinfo>BBa_K515005</partinfo>) as well as a short terminator <partinfo>BBa_B1006</partinfo> to all chromoproteins we used in this project via PCR. | ||
| + | |||
| − | |||
===Usage and Biology=== | ===Usage and Biology=== | ||
| + | The coding region of eforRed was originated from iGEM11_Uppsala-Sweden. | ||
| + | |||
| + | To achieve further control of a this reporter, we simply add a stem-loop structure(Target3) upstream of it. To activate reporter's expression, Antisense3 <partinfo>BBa_K2285010</partinfo>, which is the complementary sequence to part of Target3, is needed neither in the same plasmid or even another plasmid. | ||
| + | |||
| + | [[Image:eforRed.JPG|420px]] | ||
| + | [[Image:eforRed_tube.JPG|290px]] | ||
| + | <br>'''iGEM17_SJTU_BioX_Shanghai: '''Left picture: Chromoprotein eforRed in EP | ||
| + | <br>Right picture: ''E.coli'' with eforRed expression<br> | ||
| + | |||
| + | During experiments, we found that expression of chromoproteins amilCP <partinfo>BBa_K1357009</partinfo>, amilGFP <partinfo>BBa_K592010</partinfo>, eforRed <partinfo>BBa_K592012</partinfo> and cjBlue <partinfo>BBa_K592011</partinfo> took nearly 24 hours to see the colors with naked eyes on the LB plate. But they are still great reporters when it comes to build visualized systems which do not need to depend on extra equippments. | ||
| + | |||
| + | ===Characterization=== | ||
| + | |||
| + | In order to achieve visualization and multi-factor detection simultaneously, we selected chromoproteins as downstream expression gene. In a large number of pigment proteins, we chose those with obvious color distinction, that is the three primary colors RGB (red, green, blue) for the experiment. We used eforRed, amilGFP, cjBlue, amilCP to represent the corresponding three primary colors. | ||
| + | |||
| + | [[Image:chromoproteins.jpg|420px]] | ||
| + | |||
| + | '''Figure 2: Four kinds of chromoproteins we used in the construction of expression vector.''' | ||
| + | |||
| + | In the experiment, we linked Targets with four chromoproteins. On the dual-expression vector (STAR1 system on pCDFDuet-1 vector, STAR2 system on pETDuet-1 vector) and the relevant Antisense 1 and Antisense 3. Therefore, for future users, this kind of gene circuit can be inserted downstream of other corresponding response elements, such as operon, repressor and so on to obtain different expression of chromoproteins induced by different external stimuli, and then the visible depth of different colors can reflect the quantity of the stimuli. | ||
| + | |||
| + | [[Image:composite.jpg|420px]] | ||
| + | |||
| + | '''Figure 3: Target + chromoprotein + Antisense construct. '''In the project we inserted two genes in the same expression vector, so transforming one plasmid can achieve indiction of a certain factor. In this way, not only the burden of the host can be reduced, but the multifactorial detection will be convenient as well. | ||
| + | |||
| + | In order to prevent the limitations and errors produced by human eyes and achieve greater accuracy and convenience to measure on the basis of the visualization effect, and at the same time to meet the easy operation and cost-effective characteristics, we designed a simple pumping device, an app for analysis of results and a box providing a stable environment for the camera. More detailed information please look to the design page in our wiki. http://2017.igem.org/Team:SJTU-BioX-Shanghai/Design | ||
| + | |||
| + | [[Image:filter.jpg|420px]][[Image:box.jpg|420px]] | ||
| + | |||
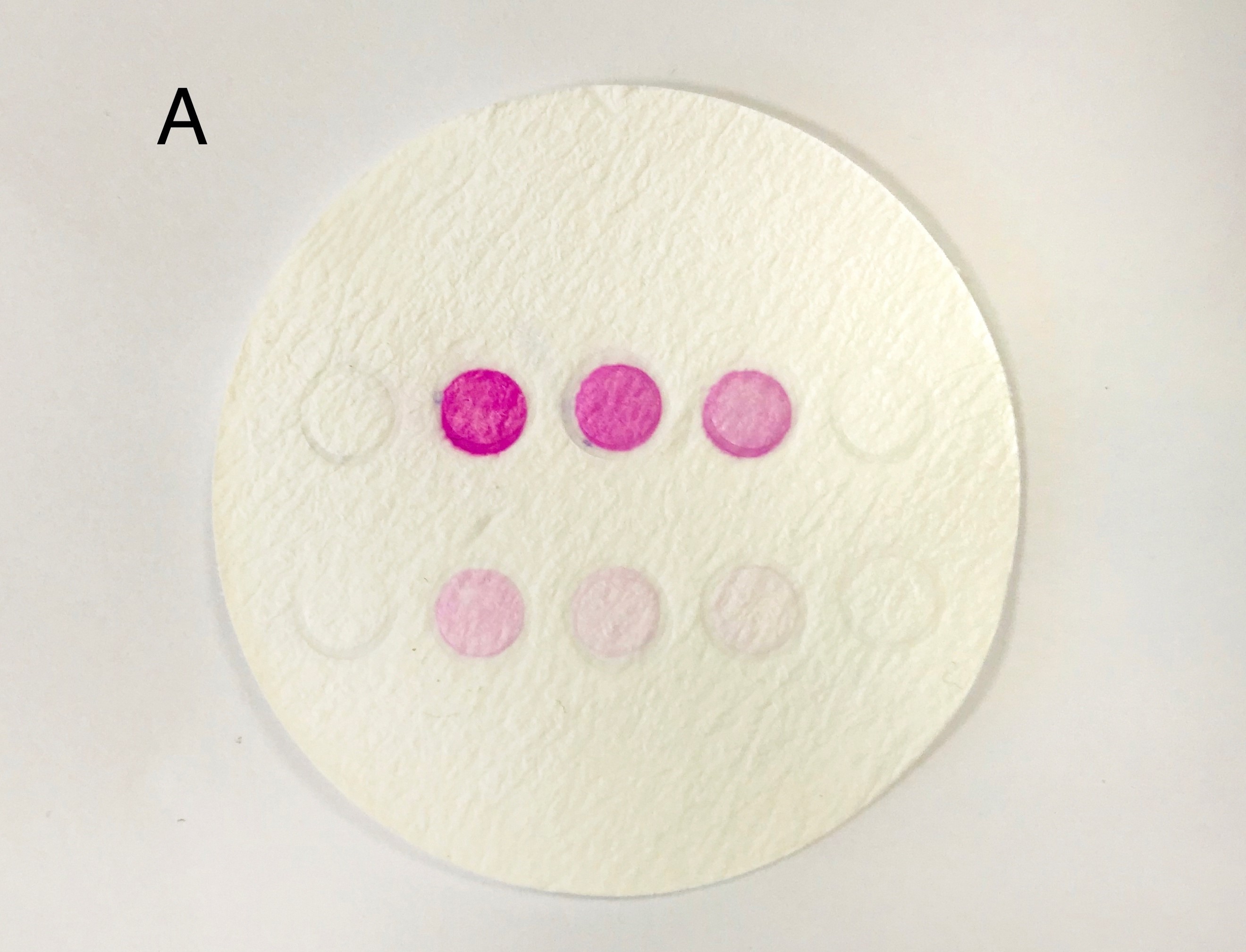
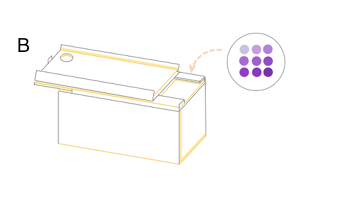
| + | '''Figure 7: the result of filtration and box and app front page '''(A) pET duet-1 plasmid with eforRed gene was transformed into BL21(DH3) E. coli, and cells with different concentration were filtered on the glass fiber filter. (B) the schematic map of the box which can give a proper distance between mobile phone and filter to be photographed. We already have a finished product of this box, to see more information please click the device page. http://2017.igem.org/Team:SJTU-BioX-Shanghai/Loader | ||
<!-- --> | <!-- --> | ||
Latest revision as of 16:19, 16 October 2018
J23119+Target3+eforRed(with RBS and Term added)
constitutive promoter J23119 followed by a stem-loop structure Target3, with chromoprotein eforRed as the reporter gene. The original part of eforRed chromoprotein is presented on BBa_K592012. What should be noticed is, the original part is a coding part and doesn't have its own RBS and Term. Therefore, to achieve greater convenience fo our Star-chromoprotein reporter system, we add the RBS which is on the upstream of sfGFP(BBa_K515005) as well as a short terminator BBa_B1006 to all chromoproteins we used in this project via PCR.
Usage and Biology
The coding region of eforRed was originated from iGEM11_Uppsala-Sweden.
To achieve further control of a this reporter, we simply add a stem-loop structure(Target3) upstream of it. To activate reporter's expression, Antisense3 BBa_K2285010, which is the complementary sequence to part of Target3, is needed neither in the same plasmid or even another plasmid.
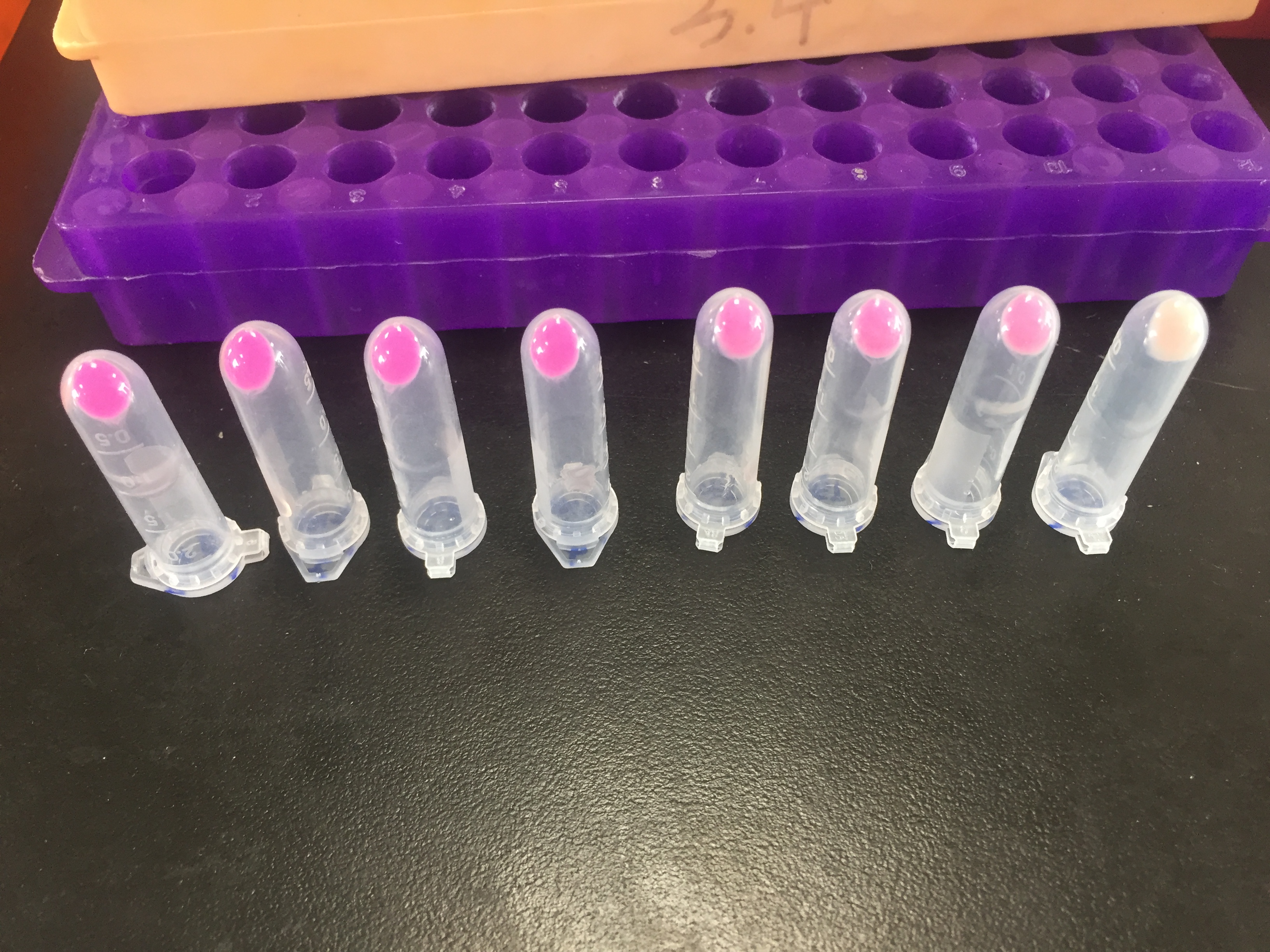

iGEM17_SJTU_BioX_Shanghai: Left picture: Chromoprotein eforRed in EP
Right picture: E.coli with eforRed expression
During experiments, we found that expression of chromoproteins amilCP BBa_K1357009, amilGFP BBa_K592010, eforRed BBa_K592012 and cjBlue BBa_K592011 took nearly 24 hours to see the colors with naked eyes on the LB plate. But they are still great reporters when it comes to build visualized systems which do not need to depend on extra equippments.
Characterization
In order to achieve visualization and multi-factor detection simultaneously, we selected chromoproteins as downstream expression gene. In a large number of pigment proteins, we chose those with obvious color distinction, that is the three primary colors RGB (red, green, blue) for the experiment. We used eforRed, amilGFP, cjBlue, amilCP to represent the corresponding three primary colors.
Figure 2: Four kinds of chromoproteins we used in the construction of expression vector.
In the experiment, we linked Targets with four chromoproteins. On the dual-expression vector (STAR1 system on pCDFDuet-1 vector, STAR2 system on pETDuet-1 vector) and the relevant Antisense 1 and Antisense 3. Therefore, for future users, this kind of gene circuit can be inserted downstream of other corresponding response elements, such as operon, repressor and so on to obtain different expression of chromoproteins induced by different external stimuli, and then the visible depth of different colors can reflect the quantity of the stimuli.
Figure 3: Target + chromoprotein + Antisense construct. In the project we inserted two genes in the same expression vector, so transforming one plasmid can achieve indiction of a certain factor. In this way, not only the burden of the host can be reduced, but the multifactorial detection will be convenient as well.
In order to prevent the limitations and errors produced by human eyes and achieve greater accuracy and convenience to measure on the basis of the visualization effect, and at the same time to meet the easy operation and cost-effective characteristics, we designed a simple pumping device, an app for analysis of results and a box providing a stable environment for the camera. More detailed information please look to the design page in our wiki. http://2017.igem.org/Team:SJTU-BioX-Shanghai/Design
Figure 7: the result of filtration and box and app front page (A) pET duet-1 plasmid with eforRed gene was transformed into BL21(DH3) E. coli, and cells with different concentration were filtered on the glass fiber filter. (B) the schematic map of the box which can give a proper distance between mobile phone and filter to be photographed. We already have a finished product of this box, to see more information please click the device page. http://2017.igem.org/Team:SJTU-BioX-Shanghai/Loader
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]