Difference between revisions of "Part:BBa K2278023"
| (28 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
| − | |||
__NOTOC__ | __NOTOC__ | ||
| − | <partinfo> | + | <partinfo>BBa_K2278023 short</partinfo> |
| − | + | Sequence and features | |
| − | <partinfo> | + | <partinfo>BBa_K2278023 SequenceAndFeatures</partinfo> |
=='''Introduction'''== | =='''Introduction'''== | ||
<html> | <html> | ||
| − | This DNA biobrick was designed in order to produce | + | This DNA biobrick was designed in order to produce cOT2 antimicrobial peptide. |
| − | + | ||
<h3 id="RT"> 1- Biological background </h3> | <h3 id="RT"> 1- Biological background </h3> | ||
| + | Antimicrobial peptides (AMP) are phylogenetically ancient components of the innate defense of both invertebrates and vertebrates. In the context of growing bacterial antibiotic-resistance, these AMP are considered as potential new therapeutical candidates. | ||
| + | <br> | ||
| + | Crocodile ovotransferrin 2 peptide (cOT2) is an engineered peptides coming from the siamese crocodile. It bears the natural sequence of cOT1 and has been extended based on the <i>C. siamensis</i> transferrin amino sequence to increase its natural antimicrobial activity The peptide is a 29 amino acid residue : KKSCHTGLKKSAGWVIPIGTLVKNGIIVR. The mechanism of action of cOT2 has been observed scanning electron microscopy. This cationic and amphipathic molecules is able to attach to and insert into membrane bilayers to form pores triggering bacterial cell lysis. | ||
| + | <br> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<h3 id="RT"> 2- Usage in iGEM projects </h3> | <h3 id="RT"> 2- Usage in iGEM projects </h3> | ||
| − | + | The part was designed during the Croc’n Cholera project <a href="http://2017.igem.org/Team:INSA-UPS_France">(team INSA-UPS-France 2017)</a>. It produces the cOT2 AMP when associated with a yeast promoter. The α-factor (</html><partinfo>BBa_K1800001</partinfo><html>) sequence contains a RBS and a signal sequence to secrete the produced peptides. | |
| − | The part was designed | + | |
| − | + | ||
| − | + | ||
| − | + | ||
</html> | </html> | ||
| + | <br> | ||
| + | <br> | ||
=='''Experiments'''== | =='''Experiments'''== | ||
| Line 39: | Line 26: | ||
<h3 id="RT"> 1- Molecular biology </h3> | <h3 id="RT"> 1- Molecular biology </h3> | ||
<p> | <p> | ||
| − | The | + | The gene was placed under the control of an alpha factor signal. IDT performed the DNA synthesis and delivered the part as gBlock. The construct was cloned by conventional ligation into the pSB1C3 plasmid. The construction was then inserted on plasmid pPICZa and integrated in the yeast genome. |
| − | + | ||
| − | The construct was cloned by conventional ligation into pSB1C3 plasmid | + | |
| − | The construction was then inserted on plasmid pPICZa and integrated in the yeast genome. | + | |
</p> | </p> | ||
| − | + | <br> | |
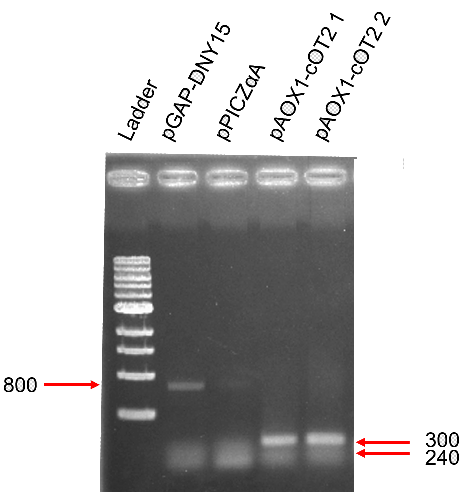
<b>Analysis of the restriction map </b> | <b>Analysis of the restriction map </b> | ||
| − | < | + | </html> |
| + | [[Image:T--INSA-UPS_France--cOT2.png|800px|thumb|center|'''Figure 1:''' <b>Analysis of the restriction map BBa_K2278023. </b> Digested fragments (XbaI and PstI) are electrophoresed through a 1% agarose gel. Control vector pSB1C3 contained an insert and expected size were 2034 and 700 bp (digestion was not total, hence the 2734 bp fragment). The fragment lengths of the correct clones (#2 to #5) were 2035 bp and 385 bp for the required insert.]] | ||
| + | <html> | ||
| + | <br> | ||
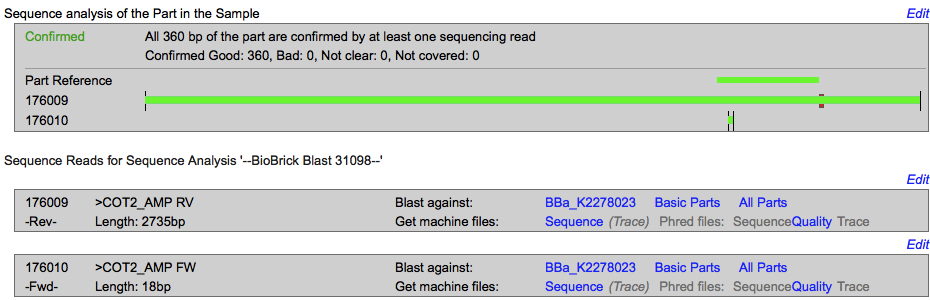
<p><b>Sequencing </p></b> | <p><b>Sequencing </p></b> | ||
| − | |||
| + | </html> | ||
| + | [[Image:T--INSA-UPS_France--coT2seq.png|800px|thumb|center|'''Figure 2:''' <b>Sequencing of pSB1C3-cOT2 </b> 1500 ng of plasmid are sequenced. The obtained sequence were blast on the BBa_K2278023 sequence with the iGEM sequencing online tools.]] | ||
| + | <html> | ||
The sequencing successfully validated the sequence of the biobrick. | The sequencing successfully validated the sequence of the biobrick. | ||
| + | <br> | ||
| + | <h3 id="RT"> 2- Integration in <i>Pichia pastoris</i> </h3> | ||
| + | The biobrick was placed under the control of the pAOXI promoter and was cloned in the pPICZalpha vector, an expression vector for the yeast <i>Pichia pastoris</i>. | ||
| + | The plasmid was then linearized and transferred in <i>Pichia pastoris</i> by electroporation. The integration is predicted to be at the pAOXI location. Indeed, the pAOXI promoter makes genome recombination easier in <i>Pichia pastoris</i>. | ||
| − | + | </html> | |
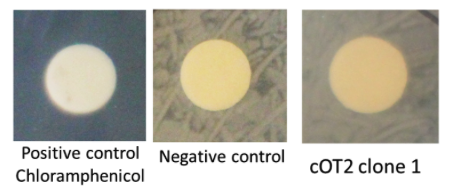
| + | [[Image:T--INSA-UPS_France--_pcrcolony_cot2.png|800px|thumb|center|'''Figure 3:''' <b>Integration of pAOXI+BBa_K2278023 in <i>Pichia pastoris</i> </b> To verify the correct integration, we performed colony PCR and electrophoresed the product through a 1% agarose gel. A 307 bp fragment is expected if the integration is correct. ]] | ||
| − | < | + | <html> |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| + | Correct amplifications were observed for the 2 colonies tested and the positive controls with the D-NY15 fragment as matrix (D-NY15). Negative control with pPICZalpha presented a non specific amplification band with a size inferior the 307 bp. | ||
</html> | </html> | ||
=='''Characterization'''== | =='''Characterization'''== | ||
<html> | <html> | ||
| + | <h3 id="RT">2. Toxicity assay </h3> | ||
| + | cOT2 production was performed with <i>Pichia pastoris</i> rown for 4 days at 30 °C with shacking in YPD 40 g/L glucose plus methanol to trigger the pAOX1 promoter. Supernatants from yeasts with or without the cOT2 encoding gene were sampled. The supernatants were used in a halo assay against <i>V. harveyi</i> as the target of cOT2. Briefly, 35mL of supernatants were freeze-dried and then resuspended in 3.5mL of water. A paper cut was soaked with one of these solutions and placed on a Petri plate inoculated with <i>V. harveyi</i> (figure 3). | ||
| − | < | + | </html> |
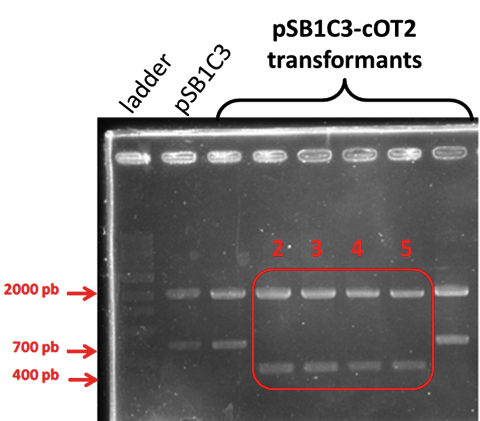
| + | [[Image:T--INSA-UPS_France--cOT2inhibition.png|800px|thumb|center|'''Figure 4:''' <b>AMP halo assay.</b>Positive control was performed with chloramphenicol (25 g/L), the negative control was performed with the empty plasmid integrated in <i>P. pastoris</i>, the assay was performed with the plasmid containing BBa_K2278023 integrated in <i>P. pastoris</i>.]] | ||
| − | + | <html> | |
| − | < | + | |
| − | No inhibition halo was observed around the yeast patch. The cOT2 cytoxicity can not be demonstrated. | + | <p><b>Conclusion : </b> </p> |
| + | No inhibition halo was observed around the yeast patch. The cOT2 cytoxicity can not be demonstrated. | ||
| + | <p><b>Perspectives: </b></p> | ||
| + | Higher concentration of yeast supernatants could be tried. | ||
| + | <br> | ||
| + | <br> | ||
| + | </html> | ||
| + | ===Design Notes=== | ||
| + | <p>Part:BBa_K1800001: Alpha-Factor Secretion Signal | ||
| + | ===Source=== | ||
| + | The peptides DNA sequence has been obtained by reverse translate the amino acid sequence of cOT2 proposed by Prajanban<i>et al</i>., 2011. They had determinated the amino acid sequence by mass spectrometry analysis. | ||
| − | + | ===References=== | |
| + | Prajanban, B., Jangpromma, N., Araki, T. and Klaynongsruang, S. (2017). Antimicrobial effects of novel peptides cOT2 and sOT2 derived from Crocodylus siamensis and Pelodiscus sinensis ovotransferrins. Biochimica et Biophysica Acta (BBA) - Biomembranes, 1859(5), pp.860-869. | ||
Latest revision as of 21:38, 23 October 2017
cOT2 antimicrobial peptide with Alpha-Factor Secretion Signal
Sequence and features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal XhoI site found at 244
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Introduction
This DNA biobrick was designed in order to produce cOT2 antimicrobial peptide.
1- Biological background
Antimicrobial peptides (AMP) are phylogenetically ancient components of the innate defense of both invertebrates and vertebrates. In the context of growing bacterial antibiotic-resistance, these AMP are considered as potential new therapeutical candidates.Crocodile ovotransferrin 2 peptide (cOT2) is an engineered peptides coming from the siamese crocodile. It bears the natural sequence of cOT1 and has been extended based on the C. siamensis transferrin amino sequence to increase its natural antimicrobial activity The peptide is a 29 amino acid residue : KKSCHTGLKKSAGWVIPIGTLVKNGIIVR. The mechanism of action of cOT2 has been observed scanning electron microscopy. This cationic and amphipathic molecules is able to attach to and insert into membrane bilayers to form pores triggering bacterial cell lysis.
2- Usage in iGEM projects
The part was designed during the Croc’n Cholera project (team INSA-UPS-France 2017). It produces the cOT2 AMP when associated with a yeast promoter. The α-factor (BBa_K1800001) sequence contains a RBS and a signal sequence to secrete the produced peptides.Experiments
1- Molecular biology
The gene was placed under the control of an alpha factor signal. IDT performed the DNA synthesis and delivered the part as gBlock. The construct was cloned by conventional ligation into the pSB1C3 plasmid. The construction was then inserted on plasmid pPICZa and integrated in the yeast genome.
Analysis of the restriction map
Sequencing
The sequencing successfully validated the sequence of the biobrick.
2- Integration in Pichia pastoris
The biobrick was placed under the control of the pAOXI promoter and was cloned in the pPICZalpha vector, an expression vector for the yeast Pichia pastoris. The plasmid was then linearized and transferred in Pichia pastoris by electroporation. The integration is predicted to be at the pAOXI location. Indeed, the pAOXI promoter makes genome recombination easier in Pichia pastoris.Correct amplifications were observed for the 2 colonies tested and the positive controls with the D-NY15 fragment as matrix (D-NY15). Negative control with pPICZalpha presented a non specific amplification band with a size inferior the 307 bp.
Characterization
2. Toxicity assay
cOT2 production was performed with Pichia pastoris rown for 4 days at 30 °C with shacking in YPD 40 g/L glucose plus methanol to trigger the pAOX1 promoter. Supernatants from yeasts with or without the cOT2 encoding gene were sampled. The supernatants were used in a halo assay against V. harveyi as the target of cOT2. Briefly, 35mL of supernatants were freeze-dried and then resuspended in 3.5mL of water. A paper cut was soaked with one of these solutions and placed on a Petri plate inoculated with V. harveyi (figure 3).
Conclusion :
No inhibition halo was observed around the yeast patch. The cOT2 cytoxicity can not be demonstrated.Perspectives:
Higher concentration of yeast supernatants could be tried.Design Notes
Part:BBa_K1800001: Alpha-Factor Secretion Signal
Source
The peptides DNA sequence has been obtained by reverse translate the amino acid sequence of cOT2 proposed by Prajanbanet al., 2011. They had determinated the amino acid sequence by mass spectrometry analysis.