Difference between revisions of "Part:BBa K1974022"
| (26 intermediate revisions by 3 users not shown) | |||
| Line 8: | Line 8: | ||
[[File:NCTU_FORMOSA_SL.png|800px|thumb|center|'''Figure 1.''' P<sub>T7</sub>+RBS+Sf1a+linker+snowdrop-lectin+linker+6X His-Tag ]] | [[File:NCTU_FORMOSA_SL.png|800px|thumb|center|'''Figure 1.''' P<sub>T7</sub>+RBS+Sf1a+linker+snowdrop-lectin+linker+6X His-Tag ]] | ||
| − | By ligating the IPTG induced P<sub>T7</sub>(BBa_ I712074), strong ribosome binding site (BBa_B0034), Sf1a, linker, snowdrop lectin ( | + | By ligating the IPTG induced P<sub>T7</sub>(BBa_ I712074), strong ribosome binding site (BBa_B0034), Sf1a, linker, snowdrop lectin (<html><a href="https://parts.igem.org/wiki/index.php?title=Part:BBa_K1974020">BBa_K1974020</a></html> |
| + | ) and the 6X His-Tag (BBa_ K1223006), we can express Sf1a, the toxin by IPTG induction. | ||
<br> | <br> | ||
This year we create a revolutionary system that integrates biological pesticides, automatic detector, sprinkler, and IoT. We made a database that contains most of the spider toxins and selected the target toxins by programming. μ-segestritoxin-Sf1a is coded for the venom of a spider, <i>Hadronyche versuta</i>.<br> | This year we create a revolutionary system that integrates biological pesticides, automatic detector, sprinkler, and IoT. We made a database that contains most of the spider toxins and selected the target toxins by programming. μ-segestritoxin-Sf1a is coded for the venom of a spider, <i>Hadronyche versuta</i>.<br> | ||
It is under the control of the strong P<sub>T7</sub>. Snowdrop-lectin acts as a carrier that could transport the toxin to insect’s nervous system, hemolymph and can improve the oral activity. A 6X His-Tag is added for further protein purification.<br> | It is under the control of the strong P<sub>T7</sub>. Snowdrop-lectin acts as a carrier that could transport the toxin to insect’s nervous system, hemolymph and can improve the oral activity. A 6X His-Tag is added for further protein purification.<br> | ||
According to reference, snowdrop-lectin is resistant to high temperature and would not be degraded by digestive juice. The species-specificity is based on the toxin, and the snowdrop lectin is the role of the carrier.<sup>[1][2]</sup> | According to reference, snowdrop-lectin is resistant to high temperature and would not be degraded by digestive juice. The species-specificity is based on the toxin, and the snowdrop lectin is the role of the carrier.<sup>[1][2]</sup> | ||
| − | + | <br> | |
<!--圖*2--> | <!--圖*2--> | ||
| Line 23: | Line 24: | ||
<p style="padding-top:20px;font-size:20px"><b>Features of Sf1a</b></p> | <p style="padding-top:20px;font-size:20px"><b>Features of Sf1a</b></p> | ||
| − | <p style="padding:1px;font-size:16px"><b>1. Non-toxic</b></p>μ-segestritoxin-Sf1a is non-toxic to mammals | + | <p style="padding:1px;font-size:16px"><b>1. Non-toxic</b></p>μ-segestritoxin-Sf1a is non-toxic to mammals. Since the structure of the target ion channel is different, μ-segestritoxin-Sf1a does not harm mammals.<sup>[4]</sup> So it is safe to use it as a biological pesticide. |
<br> | <br> | ||
<p style="padding:1px;font-size:16px"><b>2. Biodegradable</b></p>μ-segestritoxin-Sf1a is a polypeptide so it must degrade over time. After degradation, the toxin will become nutrition in the soil. | <p style="padding:1px;font-size:16px"><b>2. Biodegradable</b></p>μ-segestritoxin-Sf1a is a polypeptide so it must degrade over time. After degradation, the toxin will become nutrition in the soil. | ||
| Line 40: | Line 41: | ||
<h1>'''Experiment'''</h1> | <h1>'''Experiment'''</h1> | ||
| − | <p style="padding:1px;font-size:16px"><b>1. Cloning </b></p><br>After assembling the DNA sequences from the basic parts, we recombined each P<sub>T7</sub>+B0034+Sf1a+ linker + snowdrop -lectin +linker+ 6X His-Tag gene to pSB1C3 backbones and conducted a PCR experiment to check the size of each part. The DNA sequence length of these parts is around 600~700 b.p. In this PCR experiment, the toxin product's size should be near at 850-950 b.p.Proved that we successfully ligated the toxin sequence onto an ideal backbone. | + | <p style="padding:1px;font-size:16px"><b>1. Cloning </b></p><br>After assembling the DNA sequences from the basic parts, we recombined each P<sub>T7</sub>+B0034+Sf1a+ linker + snowdrop -lectin +linker+ 6X His-Tag gene to pSB1C3 backbones and conducted a PCR experiment to check the size of each part. The DNA sequence length of these parts is around 600~700 b.p. In this PCR experiment, the toxin product's size should be near at 850-950 b.p. Proved that we successfully ligated the toxin sequence onto an ideal backbone. |
[[File:NCTU Formosa SL-1.jpg|200px|thumb|center|'''Figure 3.''' P<sub>T7</sub> + RBS + Sf1a+linker+Lectin+linker+6X His-Tag | [[File:NCTU Formosa SL-1.jpg|200px|thumb|center|'''Figure 3.''' P<sub>T7</sub> + RBS + Sf1a+linker+Lectin+linker+6X His-Tag | ||
<br>The DNA sequence length of P<sub>T7</sub> + RBS + Sf1a+linker+6X His-Tag is around 600~700 b.p. In this PCR experiment, the product’s size should be close to 850-950 b.p.]] | <br>The DNA sequence length of P<sub>T7</sub> + RBS + Sf1a+linker+6X His-Tag is around 600~700 b.p. In this PCR experiment, the product’s size should be close to 850-950 b.p.]] | ||
| − | + | <br> | |
<!--PCR圖---> | <!--PCR圖---> | ||
| − | <p style="padding:1px;font-size:16px"><b>2. Expressing</b></p><br>We chose <i>E.coli</i> Rosetta gami strain, which can form the disulfide bonds in the cytoplasm to express the protein. To verify the <i>E.coli</i> express the P<sub>T7</sub> + RBS + Sf1a+linker+Lectin+linker+6X His-Tag which contains disulfide bonds, we treated the sample in two different ways. A means adding β-mercaptoethanol and sample buffer. β-mercaptoethanol can break the disulfide bonds of P<sub>T7</sub> + RBS + Sf1a+linker+Lectin+linker+6X His-Tag and make it a linear form. | + | <p style="padding:1px;font-size:16px"><b>2. Expressing</b></p><br>We chose <i>E. coli</i> Rosetta gami strain, which can form the disulfide bonds in the cytoplasm to express the protein. To verify the <i>E. coli</i> express the P<sub>T7</sub> + RBS + Sf1a+linker+Lectin+linker+6X His-Tag which contains disulfide bonds, we treated the sample in two different ways. A means adding β-mercaptoethanol and sample buffer. β-mercaptoethanol can break the disulfide bonds of P<sub>T7</sub> + RBS + Sf1a+linker+Lectin+linker+6X His-Tag and make it a linear form. |
The other one adding sample buffer is the native form of P<sub>T7</sub> + RBS + Sf1a+linker+Lectin+linker+6X His-Tag which maintains its structure. B is adding only sample buffer. The two samples are treated in boiling water for 15 mins. | The other one adding sample buffer is the native form of P<sub>T7</sub> + RBS + Sf1a+linker+Lectin+linker+6X His-Tag which maintains its structure. B is adding only sample buffer. The two samples are treated in boiling water for 15 mins. | ||
The SDS-PAGE shows that the native P<sub>T7</sub> + RBS+Sf1a+linker+Lectin+linker+6X His-Tag is smaller than linear one because the disulfide bonds in P<sub>T7</sub> + RBS + Sf1a+linker+Lectin+linker+6X His-Tag make the whole structure a globular shape. | The SDS-PAGE shows that the native P<sub>T7</sub> + RBS+Sf1a+linker+Lectin+linker+6X His-Tag is smaller than linear one because the disulfide bonds in P<sub>T7</sub> + RBS + Sf1a+linker+Lectin+linker+6X His-Tag make the whole structure a globular shape. | ||
| Line 58: | Line 59: | ||
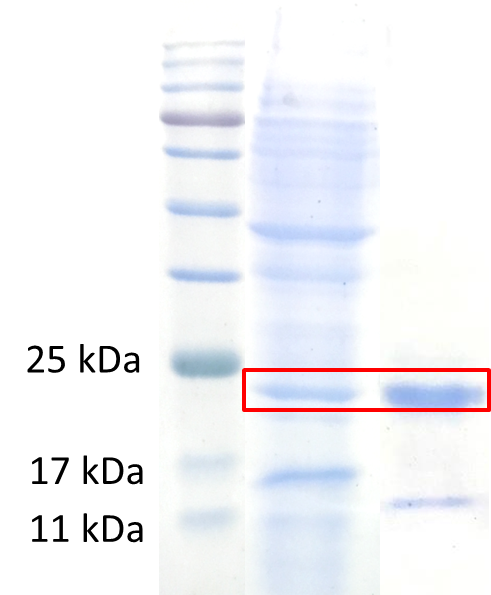
<p style="padding:1px;font-size:16px"><b>3. Purification</b></p>We sonicated the bacteria and purified the protein by 6X His-Tag behind the peptide using Nickel resin column. Then we ran the SDS-PAGE to verify the purification and analyze the concentration of P<sub>T7</sub> + RBS + Sf1a+linker+Lectin+linker+6X His-Tag. | <p style="padding:1px;font-size:16px"><b>3. Purification</b></p>We sonicated the bacteria and purified the protein by 6X His-Tag behind the peptide using Nickel resin column. Then we ran the SDS-PAGE to verify the purification and analyze the concentration of P<sub>T7</sub> + RBS + Sf1a+linker+Lectin+linker+6X His-Tag. | ||
| − | + | <br> | |
[[File:NCTU_SL-purify .png|400px|thumb|center|'''Figure 5.''' Protein electrophoresis of P<sub>T7</sub> + RBS + Sf1a+linker+Lectin+linker+6X His-Tag purification. | [[File:NCTU_SL-purify .png|400px|thumb|center|'''Figure 5.''' Protein electrophoresis of P<sub>T7</sub> + RBS + Sf1a+linker+Lectin+linker+6X His-Tag purification. | ||
<br>A is the sonication product. B is the elution product of purification.]] | <br>A is the sonication product. B is the elution product of purification.]] | ||
| Line 72: | Line 73: | ||
<!-- 放device的真實圖--> | <!-- 放device的真實圖--> | ||
<H1>'''Results'''</H1> | <H1>'''Results'''</H1> | ||
| − | Pantide-expressed E. coli Rosetta gami strain and diluted it with the three concentration.We applied the sample onto the leaf disks and put five cutworms into the separate cabinets for feeding assays. The positive control in the experiment was to apply Bacillus thuringiensis, which is the most widely-used bioinsecticide. We preserved all the result of the remained leaves sealing with the glass paper and calculated the ratio of the remained area on the leaves. The collected data were analyzed by t – test. Here are the feeding assay results. | + | Pantide-expressed <i>E. coli</i> Rosetta gami strain and diluted it with the three concentration.We applied the sample onto the leaf disks and put five cutworms into the separate cabinets for feeding assays. The positive control in the experiment was to apply <i>Bacillus thuringiensis</i>, which is the most widely-used bioinsecticide. We preserved all the result of the remained leaves sealing with the glass paper and calculated the ratio of the remained area on the leaves. The collected data were analyzed by t – test. Here are the feeding assay results. |
| − | [[File:NCTU_DOSE_SL_1.png|400px|thumb|center|'''Figure 6.''' Above is leaves remaining area of Negative control ( | + | [[File:NCTU_DOSE_SL_1.png|400px|thumb|center|'''Figure 6.''' Above is leaves remaining area of Negative control ( DDH<sub>2</sub>O ), Positive control ( <i>Bacillus thuringiensis</i> bacteria ), Sf1a+linker+6XHis-Tag, Sf1a+linker+snowdrop-lectin+linker+6XHis-Tag]] |
| − | [[File:NCTU_leaves_sl_1.png|400px|thumb|center|'''Figure 7.''' Above are leaves with of Negative control ( | + | [[File:NCTU_leaves_sl_1.png|400px|thumb|center|'''Figure 7.''' Above are leaves with of Negative control ( DDH<sub>2</sub>O ), Positive control ( <i>Bacillus thuringiensis</i> bacteria ), Sf1a+linker+snowdrop-lectin+linker+6XHis-Tag]] |
<h1>'''Safety'''</h1> | <h1>'''Safety'''</h1> | ||
| − | <p style="padding:1px;">We | + | <p style="padding:1px;"> |
| − | + | We do the safety part which is the same as the Hv1a-lectin. See more in | |
| − | + | <html><a href="https://parts.igem.org/wiki/index.php?title=Part:BBa_K1974021">BBa_K1974021</a></html>. | |
<h1>'''Reference'''</h1> | <h1>'''Reference'''</h1> | ||
| Line 98: | Line 99: | ||
<partinfo>BBa_K1974022 parameters</partinfo> | <partinfo>BBa_K1974022 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | |||
| + | : (links to uploads relevant to your contribution) | ||
| + | |||
| + | <h1>'''Contribution'''</h1> | ||
| + | <br> | ||
| + | <b> Group: [http://2018.igem.org/Team:NCTU_Formosa NCTU_Formosa 2018] </b> | ||
| + | <br> | ||
| + | <b> Author: Chen, Yen-Ling</b> | ||
| + | |||
| + | |||
| + | <h3>New improved part | ||
| + | <html><a href="https://parts.igem.org/Part:BBa_K2599016">BBa_K2599016</a></html> | ||
| + | </h3> | ||
| + | |||
| + | To enhance the function of fusion proteins and provide a proper folding of proteins, | ||
| + | NCTU_Formosa 2018 modified the linker between Sf1a (spider toxin) and lectin (orally active protein) by elongating short linker AAA (3 a.a.) to GS linker (18 a.a.). | ||
| + | |||
| + | |||
| + | The improvement part that NCTU_Formosa 2018 modified contains the IPTG induced <sub>p</sub>T7 [https://parts.igem.org/Part:BBa_I712074 (BBa_I712074)], strong ribosome binding site [https://parts.igem.org/Part:BBa_B0034 (BBa_B0034)], Sf1a, GS linker [https://parts.igem.org/Part:BBa_K1974030 (BBa_K1974030)], snowdrop lectin [https://parts.igem.org/Part:BBa_K1974020 (BBa_K1974020)] and the 6X His-Tag [https://parts.igem.org/Part:BBa_K1223006 (BBa_K1223006)]. | ||
| + | |||
| + | |||
| + | |||
| + | <p style="padding-top:20px;font-size:25px"><b> Experiment</b></p> | ||
| + | |||
| + | <p style="padding-top:20px;font-size:20px"><b> Preparation of Bio-insecticidal Proteins</b></p> | ||
| + | |||
| + | We utilized Rosetta-gami DE3 strain to express both the previous part and improvement part. The proteins that produced was then coated on leaves respectively and each leaf was placed inside containers with same number of larvae. The leaf remaining area was observed as shown as below. | ||
| + | |||
| + | |||
| + | <p style="padding-top:20px;font-size:20px"><b> Result</b></p> | ||
| + | |||
| + | ===Expression=== | ||
| + | |||
| + | |||
| + | ===Comparison of Plant Protecting Effect of Different Design of Linker between Protein Sf1a and Lectin=== | ||
| + | |||
| + | <br> | ||
| + | |||
| + | {{#tag:html|<img style="width: 60%; padding-left: 18%;" src="https://static.igem.org/mediawiki/2018/4/4c/T--NCTU_Formosa--sf1a_fig1.png" alt="" />}} | ||
| + | <div style="width:60%; padding-left: 18%;"><p style="padding-top: 12px; font-size: 10px; text-align: center;"><b>Figure 5.</b> Comparison of plant protecting effects by changing the linker between protein Sf1a and lectin. (I): link proteins with GS linker (improvement part BBa_K2599016 ); (II): link proteins with AAA linker (previous part BBa_K1974022); (III): negative control group of Rosetta gami DE3 solution. After feeding for 7 hours, percentage of remained leaf area : improvement part > previous part > negative control.</p></div> | ||
| + | <br> | ||
| + | |||
| + | |||
| + | In conclusion, through comparison, improvement group (I) is more effective in protecting leaf from larvae consuming than previous part (II) and inferred that elongation of linker can enhance the function of fusion proteins and provide a proper folding of proteins. | ||
Latest revision as of 14:36, 17 October 2018
T7Promoter+RBS+Sf1a+linker+snowdrop-lectin+linker+6X His-Tag
Introduction:
By ligating the IPTG induced PT7(BBa_ I712074), strong ribosome binding site (BBa_B0034), Sf1a, linker, snowdrop lectin (BBa_K1974020
) and the 6X His-Tag (BBa_ K1223006), we can express Sf1a, the toxin by IPTG induction.
This year we create a revolutionary system that integrates biological pesticides, automatic detector, sprinkler, and IoT. We made a database that contains most of the spider toxins and selected the target toxins by programming. μ-segestritoxin-Sf1a is coded for the venom of a spider, Hadronyche versuta.
It is under the control of the strong PT7. Snowdrop-lectin acts as a carrier that could transport the toxin to insect’s nervous system, hemolymph and can improve the oral activity. A 6X His-Tag is added for further protein purification.
According to reference, snowdrop-lectin is resistant to high temperature and would not be degraded by digestive juice. The species-specificity is based on the toxin, and the snowdrop lectin is the role of the carrier.[1][2]
Mechanism of Sf1a
According to reference, μ-segestritoxin-Sf1a has a structure called ICK(inhibitor cysteine knot).This kind of structure contains three disulfide bonds and beta-sheet. With this structure, μ-segestritoxin-Sf1a can resist the high temperature, acid-base solution and the digest juice of insect gut. μ-segestritoxin-Sf1a can bind on insect voltage-gated Sodium channels in the central nervous system, making it paralyze and die eventually.[3]
Features of Sf1a
1. Non-toxic
μ-segestritoxin-Sf1a is non-toxic to mammals. Since the structure of the target ion channel is different, μ-segestritoxin-Sf1a does not harm mammals.[4] So it is safe to use it as a biological pesticide.
2. Biodegradable
μ-segestritoxin-Sf1a is a polypeptide so it must degrade over time. After degradation, the toxin will become nutrition in the soil.
3. Species-specific
According to reference, μ-segestritoxin-Sf1a has specificity to Lepidopteran (moths), Dipteran (flies) and Orthopteran (grasshoppers).
4. Eco-friendly
Compare with chemical pesticides, μ-segestritoxin-Sf1a will not remain in soil and water so that it will not pollute the environment and won’t harm the ecosystem.Together, using μ-segestritoxin-Sf1a is totally an environmentally friendly way for solving harmful insect problems by using this ion channel inhibitor as a biological pesticide.
Target insect:
Experiment
1. Cloning
After assembling the DNA sequences from the basic parts, we recombined each PT7+B0034+Sf1a+ linker + snowdrop -lectin +linker+ 6X His-Tag gene to pSB1C3 backbones and conducted a PCR experiment to check the size of each part. The DNA sequence length of these parts is around 600~700 b.p. In this PCR experiment, the toxin product's size should be near at 850-950 b.p. Proved that we successfully ligated the toxin sequence onto an ideal backbone.
2. Expressing
We chose E. coli Rosetta gami strain, which can form the disulfide bonds in the cytoplasm to express the protein. To verify the E. coli express the PT7 + RBS + Sf1a+linker+Lectin+linker+6X His-Tag which contains disulfide bonds, we treated the sample in two different ways. A means adding β-mercaptoethanol and sample buffer. β-mercaptoethanol can break the disulfide bonds of PT7 + RBS + Sf1a+linker+Lectin+linker+6X His-Tag and make it a linear form.
The other one adding sample buffer is the native form of PT7 + RBS + Sf1a+linker+Lectin+linker+6X His-Tag which maintains its structure. B is adding only sample buffer. The two samples are treated in boiling water for 15 mins. The SDS-PAGE shows that the native PT7 + RBS+Sf1a+linker+Lectin+linker+6X His-Tag is smaller than linear one because the disulfide bonds in PT7 + RBS + Sf1a+linker+Lectin+linker+6X His-Tag make the whole structure a globular shape.
3. Purification
We sonicated the bacteria and purified the protein by 6X His-Tag behind the peptide using Nickel resin column. Then we ran the SDS-PAGE to verify the purification and analyze the concentration of PT7 + RBS + Sf1a+linker+Lectin+linker+6X His-Tag.
4.Modeling
According to reference, the energy of Ultraviolet will break the disulfide bonds and the toxicity is also decreased. To take the parameter into consideration for our automatic system, we modeled the degradation rate of the protein and modify the program in our device.
5. Device
We designed a device that contains detector, sprinkler, and integrated hardware with users by APP through IoT talk. We use infrared detector to detect the number of the pest and predict what time to spray the farmland. Furthermore, other detectors like temperature, humidity, lamination, pressure of carbon dioxide and on also install in our device. At the same time, the APP would contact the users that all the information about the farmland and spray biological pesticides automatically. This device can make farmers control the farmland remotely.
Results
Pantide-expressed E. coli Rosetta gami strain and diluted it with the three concentration.We applied the sample onto the leaf disks and put five cutworms into the separate cabinets for feeding assays. The positive control in the experiment was to apply Bacillus thuringiensis, which is the most widely-used bioinsecticide. We preserved all the result of the remained leaves sealing with the glass paper and calculated the ratio of the remained area on the leaves. The collected data were analyzed by t – test. Here are the feeding assay results.
Safety
We do the safety part which is the same as the Hv1a-lectin. See more in BBa_K1974021.
Reference
1. Elaine Fitches, Martin G. Edwards, Christopher Mee, Eugene Grishin, Angharad M. R. Gatehouse, John P. Edwards, John A. Gatehouse “Fusion proteins containing insect-specific toxins as pest control agents: snowdrop lectin delivers fused insecticidal spider venom toxin to insect haemolymph following oral ingestion,” Journal of Insect Physiology, 2004, 50, pp.61-71
2. Elaine C. Fitches, Prashant Pyati, Glenn F. King, John A. Gatehouse, “ Fusion to Snowdrop Lectin Magnifies the Oral Activity of Insecticidal Omega-Hexatoxin-Hv1a Peptide by Enabling Its Delivery to the Central Nervous System,”
3. Monique J. Windley, Volker Herzig, Slawomir A. Dziemborowicz, Margaret C. Hardy, Glenn F. King and Graham M. Nicholson, “Spider-Venom Peptide as Bioinsecticide,” Toxins Review, 2012, 4, pp. 191-227.
4. A. Lipkin, S. Kozlov, E. Nosyreva, A. Blake, J.D. Windass, E. Grishin (2001, April 9). Novel insecticidal toxins from the venom of the spider Segestria florentina. Toxicon, 40, 125-130.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
- (links to uploads relevant to your contribution)
Contribution
Group: [http://2018.igem.org/Team:NCTU_Formosa NCTU_Formosa 2018]
Author: Chen, Yen-Ling
New improved part BBa_K2599016
To enhance the function of fusion proteins and provide a proper folding of proteins, NCTU_Formosa 2018 modified the linker between Sf1a (spider toxin) and lectin (orally active protein) by elongating short linker AAA (3 a.a.) to GS linker (18 a.a.).
The improvement part that NCTU_Formosa 2018 modified contains the IPTG induced pT7 (BBa_I712074), strong ribosome binding site (BBa_B0034), Sf1a, GS linker (BBa_K1974030), snowdrop lectin (BBa_K1974020) and the 6X His-Tag (BBa_K1223006).
Preparation of Bio-insecticidal Proteins
We utilized Rosetta-gami DE3 strain to express both the previous part and improvement part. The proteins that produced was then coated on leaves respectively and each leaf was placed inside containers with same number of larvae. The leaf remaining area was observed as shown as below.
Result
Expression
Comparison of Plant Protecting Effect of Different Design of Linker between Protein Sf1a and Lectin

Figure 5. Comparison of plant protecting effects by changing the linker between protein Sf1a and lectin. (I): link proteins with GS linker (improvement part BBa_K2599016 ); (II): link proteins with AAA linker (previous part BBa_K1974022); (III): negative control group of Rosetta gami DE3 solution. After feeding for 7 hours, percentage of remained leaf area : improvement part > previous part > negative control.
In conclusion, through comparison, improvement group (I) is more effective in protecting leaf from larvae consuming than previous part (II) and inferred that elongation of linker can enhance the function of fusion proteins and provide a proper folding of proteins.