Difference between revisions of "Part:BBa K1893016"
(→Usage and Biology) |
(→Characterisation data) |
||
| (One intermediate revision by the same user not shown) | |||
| Line 7: | Line 7: | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| − | The T7 phage is a bacteriophage that infects Escherichia coli and leads to cell lysis of the host. It is known in synthetic biology as the source of the T7 promoter, which allows for tight control of gene expression in the presence of T7 RNA polymerase. The T7 phage infection mechanism is facilitated by a number of viral genes encoded in the 40kb T7 phage genome, including gene product 2. | + | The T7 phage is a bacteriophage that infects <i>Escherichia coli</i> and leads to cell lysis of the host. It is known in synthetic biology as the source of the T7 promoter, which allows for tight control of gene expression in the presence of T7 RNA polymerase. The T7 phage infection mechanism is facilitated by a number of viral genes encoded in the 40kb T7 phage genome, including gene product 2. |
| − | Gene product 2 (gp2) is a small 7 kDA protein that plays a key role in the late stages of T7 phage infection. It binds to the β’ subunit of RNA polymerase (RNAP) in the E. coli host, which inhibits host transcription by preventing formation of the active RNAP holoenzyme. This allows the phage-encoded RNAP to transcribe the phage proteins required for successful infection without interference from the host transcriptional machinery. One effect of inhibited host transcription is a decrease in the growth rate of the host. | + | Gene product 2 (gp2) is a small 7 kDA protein that plays a key role in the late stages of T7 phage infection. It binds to the β’ subunit of RNA polymerase (RNAP) in the <i>E. coli</i> host, which inhibits host transcription by preventing formation of the active RNAP holoenzyme. This allows the phage-encoded RNAP to transcribe the phage proteins required for successful infection without interference from the host transcriptional machinery. One effect of inhibited host transcription is a decrease in the growth rate of the host. |
This part can arrest the growth of <i>E. coli</i> within half an hour of induction by arabinose. Once the pBAD promoter is switched off with glucose, the cells can recover back to normal growth rate within approximately 50 minutes. We have submitted this part as our best composite, because it represents an effective an novel way to control the size of a bacterial population, and has several advantages over existing population control methods. | This part can arrest the growth of <i>E. coli</i> within half an hour of induction by arabinose. Once the pBAD promoter is switched off with glucose, the cells can recover back to normal growth rate within approximately 50 minutes. We have submitted this part as our best composite, because it represents an effective an novel way to control the size of a bacterial population, and has several advantages over existing population control methods. | ||
| Line 19: | Line 19: | ||
===Characterisation data=== | ===Characterisation data=== | ||
[[File:Gp2.png|700px|center|]] | [[File:Gp2.png|700px|center|]] | ||
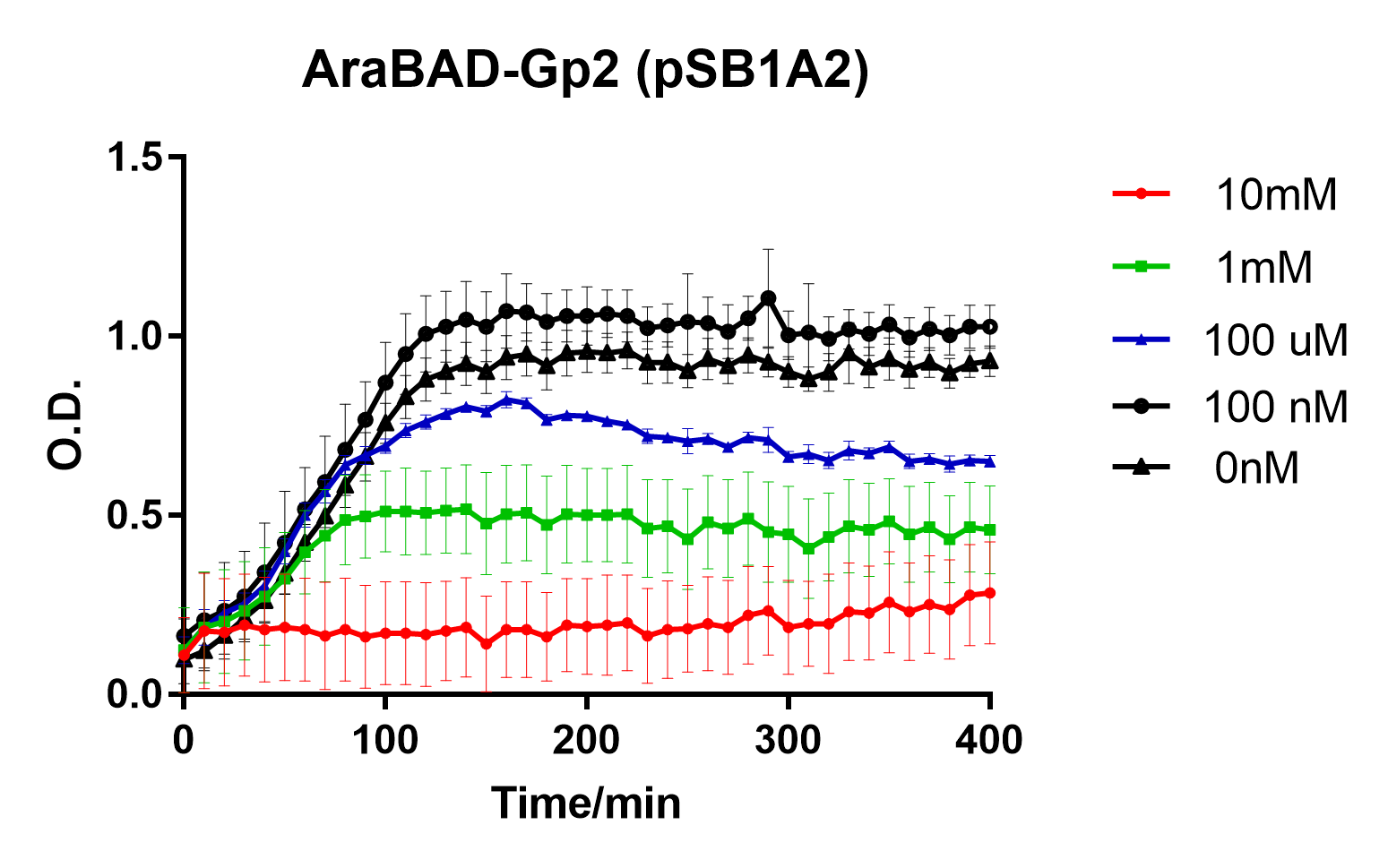
| − | Figure 1: Growth inhibition of Top10 cells by induction of Gp2 by L-arabinose. Experiments were performed in E. coli Top10 cell strain cultured at 37°C, which were diluted to 0.05 O.D. and inoculated with L-arabinose at the 0 minute timepoint. O.D. was recorded at 600nm. Reported values represent the mean normalised O.D. for three repeats, with error bars representing standard deviation. | + | Figure 1: Growth inhibition of Top10 cells by induction of Gp2 by L-arabinose. Experiments were performed in <i>E. coli Top10</i> cell strain cultured at 37°C, which were diluted to 0.05 O.D. and inoculated with L-arabinose at the 0 minute timepoint. O.D. was recorded at 600nm. Reported values represent the mean normalised O.D. for three repeats, with error bars representing standard deviation. |
[[File:Gp2_recovery.png|700px|center|]] | [[File:Gp2_recovery.png|700px|center|]] | ||
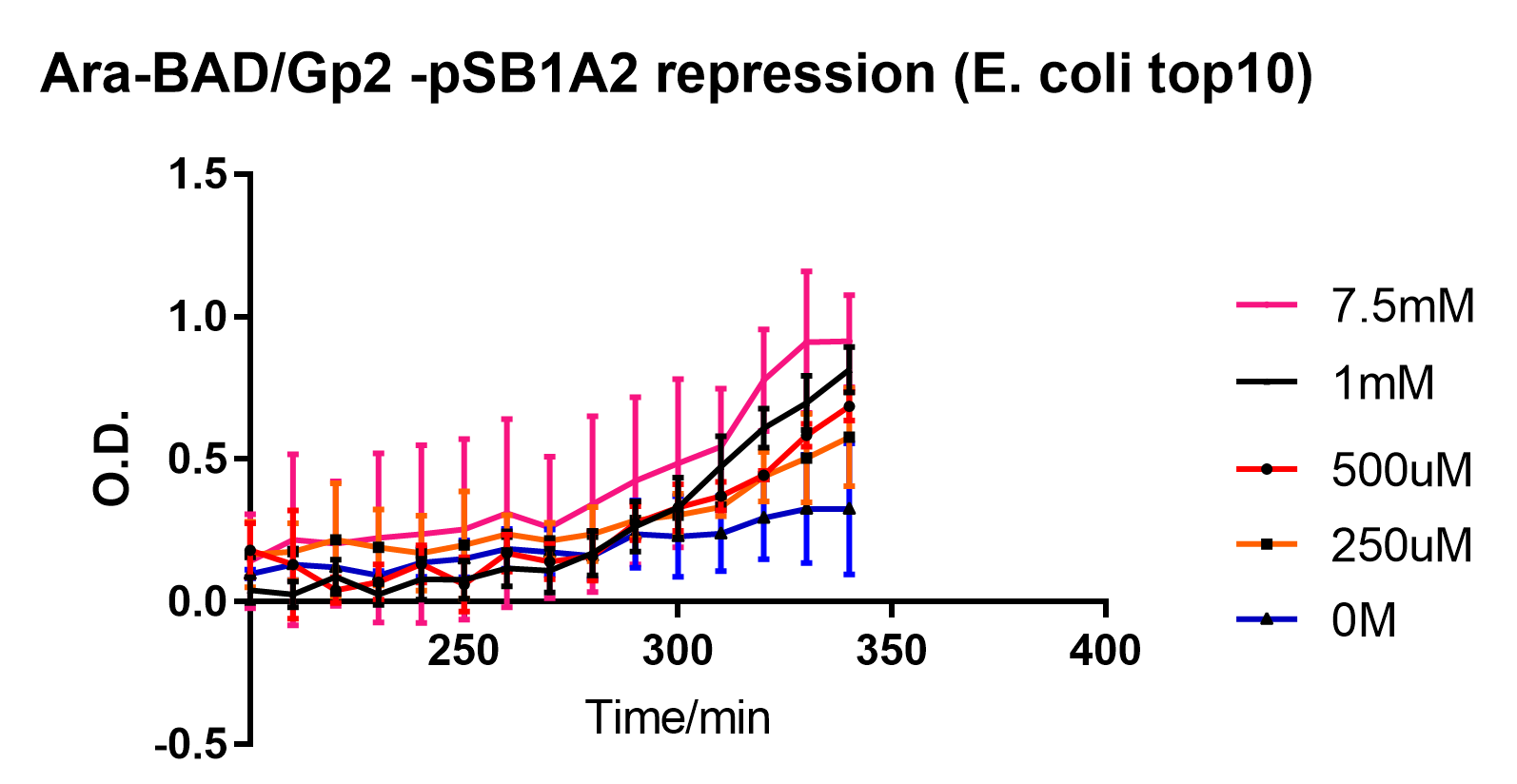
| − | Figure 2: Recovery of growth by TOP10 cells after arabinose-induced pBAD operon was switched off by glucose-mediated catabolite repression. 100uM of L-arabinose was added to the culture 2 hours before the 0 minute timepoint and D-glucose was added at the 0 minute timepoint | + | Figure 2: Recovery of growth by TOP10 cells after arabinose-induced pBAD operon was switched off by glucose-mediated catabolite repression. 100uM of L-arabinose was added to the culture 2 hours before the 0 minute timepoint and D-glucose was added at the 0 minute timepoint. pBAD-GFP controls indicate that the pBAD operon was switched off at approximately after approximately 250 minutes. Experiments were performed in E. coli Top10 cell strain cultured at 37°C, which were diluted to 0.05 O.D., which was recorded at 600nm. Reported values represent the mean normalised O.D. for three repeats, with error bars representing standard deviation. |
Latest revision as of 16:36, 28 October 2016
Arabinose inducible gp2 (pBAD+gp2)
The T7 phage gene Gp2 under control of the pBAD promoter. This part allows for robust growth inhibition in E. coli within half an hour upon addition of arabinose to the culture. Addition of glucose switches off the production of Gp2, and allows recovery of normal growth rates after several hours.
Usage and Biology
The T7 phage is a bacteriophage that infects Escherichia coli and leads to cell lysis of the host. It is known in synthetic biology as the source of the T7 promoter, which allows for tight control of gene expression in the presence of T7 RNA polymerase. The T7 phage infection mechanism is facilitated by a number of viral genes encoded in the 40kb T7 phage genome, including gene product 2.
Gene product 2 (gp2) is a small 7 kDA protein that plays a key role in the late stages of T7 phage infection. It binds to the β’ subunit of RNA polymerase (RNAP) in the E. coli host, which inhibits host transcription by preventing formation of the active RNAP holoenzyme. This allows the phage-encoded RNAP to transcribe the phage proteins required for successful infection without interference from the host transcriptional machinery. One effect of inhibited host transcription is a decrease in the growth rate of the host.
This part can arrest the growth of E. coli within half an hour of induction by arabinose. Once the pBAD promoter is switched off with glucose, the cells can recover back to normal growth rate within approximately 50 minutes. We have submitted this part as our best composite, because it represents an effective an novel way to control the size of a bacterial population, and has several advantages over existing population control methods.
Characterisation data
Figure 1: Growth inhibition of Top10 cells by induction of Gp2 by L-arabinose. Experiments were performed in E. coli Top10 cell strain cultured at 37°C, which were diluted to 0.05 O.D. and inoculated with L-arabinose at the 0 minute timepoint. O.D. was recorded at 600nm. Reported values represent the mean normalised O.D. for three repeats, with error bars representing standard deviation.
Figure 2: Recovery of growth by TOP10 cells after arabinose-induced pBAD operon was switched off by glucose-mediated catabolite repression. 100uM of L-arabinose was added to the culture 2 hours before the 0 minute timepoint and D-glucose was added at the 0 minute timepoint. pBAD-GFP controls indicate that the pBAD operon was switched off at approximately after approximately 250 minutes. Experiments were performed in E. coli Top10 cell strain cultured at 37°C, which were diluted to 0.05 O.D., which was recorded at 600nm. Reported values represent the mean normalised O.D. for three repeats, with error bars representing standard deviation.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1342
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 1281
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 1116
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 1473
Illegal SapI site found at 1098