Difference between revisions of "Part:BBa K1980011"
(→Composite) |
|||
| (7 intermediate revisions by the same user not shown) | |||
| Line 4: | Line 4: | ||
==Description== | ==Description== | ||
| − | <p> This composite part contains the cytoplasmic chelator MymT from <i>M. tuberculosis</i> behind the pCopA promoter with divergently expressed CueR (a copper sensitive transcription factor) | + | <html> |
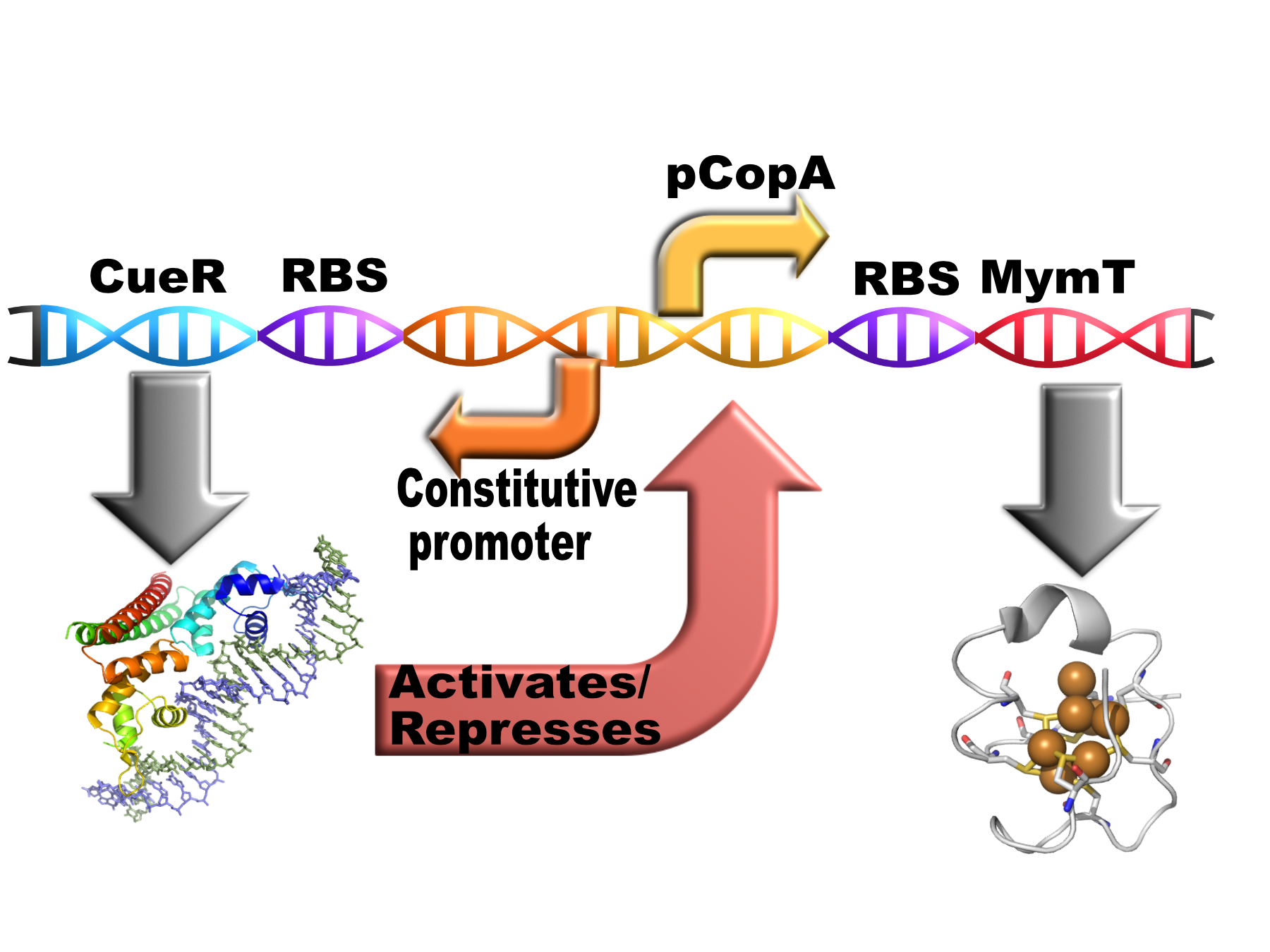
| + | <p> This composite part contains the cytoplasmic copper chelator MymT (<a href="https://parts.igem.org/Part:BBa_K1980002">BBa_K1980002</a>)from <i>M. tuberculosis</i> behind the pCopA promoter with divergently expressed CueR (a copper sensitive transcription factor)(<a href="https://parts.igem.org/Part:BBa_K1980006">BBa_K1980006</a>).</p></html> | ||
<!-- --> | <!-- --> | ||
| − | <span class='h3bb'>Sequence and Features</span> | + | <span class='h3bb'>Sequence and Features:</span> |
<partinfo>BBa_K1980011 SequenceAndFeatures</partinfo> | <partinfo>BBa_K1980011 SequenceAndFeatures</partinfo> | ||
| − | |||
==Usage and Biology== | ==Usage and Biology== | ||
| + | Our project was to investigate a probiotic treatment for the copper-accumulation disorder: Wilson's disease. This required a system able to detect dietary copper, ideally in the range over which copper concentration changes after a meal (around 5-10μM). When copper is detected a copper chelating protein should be induced to lower the free copper concentration to prevent its absorption by the body. | ||
| + | ===CueR and pCopA=== | ||
<p><i>E. coli</i> cells use a protein called CueR to regulate the cytoplasmic copper concentration. CueR is a MerR-type regulator with an interesting mechanism of action whereby it can behave as a net activator or a net repressor under different copper concentrations through interaction with RNA polymerase<sup>(1)</sup>. CueR forms dimers consisting of three functional domains (a DNA-binding, a dimerisation and a metal-binding domain). The DNA binding domains bind to DNA inverted repeats called CueR boxes with the sequence:</p> | <p><i>E. coli</i> cells use a protein called CueR to regulate the cytoplasmic copper concentration. CueR is a MerR-type regulator with an interesting mechanism of action whereby it can behave as a net activator or a net repressor under different copper concentrations through interaction with RNA polymerase<sup>(1)</sup>. CueR forms dimers consisting of three functional domains (a DNA-binding, a dimerisation and a metal-binding domain). The DNA binding domains bind to DNA inverted repeats called CueR boxes with the sequence:</p> | ||
| − | + | CCTTCCNNNNNNGGAAGG | |
| − | + | ||
<p>This box is present at the promoter regions of the copper exporting ATPase CopA, some molybdenum cofactor synthesis genes and the periplasmic copper oxidase protein CueO.<sup>(2)</sup></p> | <p>This box is present at the promoter regions of the copper exporting ATPase CopA, some molybdenum cofactor synthesis genes and the periplasmic copper oxidase protein CueO.<sup>(2)</sup></p> | ||
| + | ==MymT== | ||
| + | |||
| + | MymT is a small prokaryotic metallothein discovered in Mycobacterium tuberculosis by Gold et al.(3). It is believed that the protein may help the bacterium survive copper toxicity. | ||
| + | MymT is believed to bind up to 7 copper ions but has a preference for 4-6.(3) | ||
| + | |||
| + | ===Composite=== | ||
| + | |||
| + | <p> This is how the part is intended to work <i>in vivo</i>:</p> | ||
| + | |||
| + | [[Image:11_schematic.jpeg|400px|thumb|center|Schematic of BBa K1980011 using a yeast metallothein (PDB: 1AQR) to represent MymT]] | ||
| + | |||
| + | <p>Copper should induce the expression of the chelator via CueR. The chelator should reduce the free copper concentration therefore reducing it level of expression. This should in theory result in the free copper concentration falling to a low, stable level. </p> | ||
==Experience== | ==Experience== | ||
<p>We cloned pCopA MymT with divergent expressed CueR from a gBlock into the shipping plasmid pSB1C3. <i>E. coli</i> strain MG1655 was transformed using the specific recombinant plasmid and a 5ml culture of a transformed colony was grown overnight. </p> | <p>We cloned pCopA MymT with divergent expressed CueR from a gBlock into the shipping plasmid pSB1C3. <i>E. coli</i> strain MG1655 was transformed using the specific recombinant plasmid and a 5ml culture of a transformed colony was grown overnight. </p> | ||
| − | |||
| − | + | ===Absorbance Assay=== | |
| + | |||
| + | <p> Separately MymT was cloned from Gblock (<html><a href="https://parts.igem.org/Part:BBa_K1980002">BBa_K1980002</a></html>) into the shipping vector then transferred it into the pBAD, arabinose-inducible commercial expression vector. </p> | ||
| + | |||
| + | <p>We were unable however to detect a drop in solution copper concentration due to copper chelation activity of MymT when expressed from pBAD in MG1655 <i>E. coli</i> strain, using an absorbance assay. Performing a similar procedure with this part also showed no measurable drop in copper concentration. | ||
| + | </p> | ||
| + | <p> | ||
| + | The assay used was the reagent Bathocuproine disulphonic Acid (BCS). BCS is colourless in the absence of Cu<sup>+</sup> but upon exposure to Cu<sup>+</sup>, BCS forms complexes with Cu<sup>+</sup> and absorbs strongly in the 480nm range. In our assays, at concentrations of 50µg/mL, the 480nm absorbance varied linearly with the Cu+ concentration from the detection limit of around 1µM Cu<sup>+</sup>, to approximately 20µM Cu<sup>+</sup>. As not only MymT and Csp1 bind copper in the Cu<sup>+</sup> form, but the assay also requires singly charged copper, the assays were optimised to include a suitable about of mild reducing agent to ensure reduction of the added CuSO<sub>4</sub> (releases Cu<sup>2+</sup>) to Cu<sup>+</sup>. After trying L(+)-Ascorbate and DL-Dithiothreitol, and L-Glutathione as candidates, L-Glutathione was selected as it was both mild enough to not damage biological material and efficient at reducing Cu<sup>2+</sup>. At >2-3 times the concentration of Cu<sup>2+</sup> in solution, L-Glutathione had maximum reductive activity against Cu<sup>2+</sup>.</p> | ||
| + | <p> Modelling by our team suggested that this was because insufficient protein could be expressed to chelate the amount needed to be detectable on the assay (1μM detection limit).</p> | ||
| + | ===Conclusions=== | ||
| + | <p>We were unable to demonstrate the ability of this system to buffer free copper concentration we believe that this is because the range of concentrations over which the system operates is below the detection limit of our absorbance assay. Future work should be done to optimise the system so it can operate over a wider range of copper concentrations. This could possibly be done by changing the promoter sequence or the RBS strengths.</p> | ||
==References== | ==References== | ||
| Line 30: | Line 53: | ||
<p>(2) Yamamoto K, Ishihama A. (2005) “Transcriptional response of <i>Escherichia coli</i> to external copper.” Mol Microbiol. 2005 Apr;56(1):215-27</p> | <p>(2) Yamamoto K, Ishihama A. (2005) “Transcriptional response of <i>Escherichia coli</i> to external copper.” Mol Microbiol. 2005 Apr;56(1):215-27</p> | ||
| + | <p>(3) Ben Gold, Haiteng Deng, Ruslana Bryk, Diana Vargas, David Eliezer, Julia Roberts, Xiuju Jiang, & Carl Nathan (2009) “Identification of a Copper-Binding Metallothionein in Pathogenic Mycobacteria” Nat Chem Biol. 2008 October ; 4(10): 609–616. doi:10.1038/nchembio.109.</p> | ||
<p style="text-align: right"><i>Author: Andreas Hadjicharalambous</i></p> | <p style="text-align: right"><i>Author: Andreas Hadjicharalambous</i></p> | ||
Latest revision as of 21:25, 24 October 2016
pCopA MymT with divergent expressed CueR
Description
This composite part contains the cytoplasmic copper chelator MymT (BBa_K1980002)from M. tuberculosis behind the pCopA promoter with divergently expressed CueR (a copper sensitive transcription factor)(BBa_K1980006).
Sequence and Features:
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 438
Illegal NheI site found at 461 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Usage and Biology
Our project was to investigate a probiotic treatment for the copper-accumulation disorder: Wilson's disease. This required a system able to detect dietary copper, ideally in the range over which copper concentration changes after a meal (around 5-10μM). When copper is detected a copper chelating protein should be induced to lower the free copper concentration to prevent its absorption by the body.
CueR and pCopA
E. coli cells use a protein called CueR to regulate the cytoplasmic copper concentration. CueR is a MerR-type regulator with an interesting mechanism of action whereby it can behave as a net activator or a net repressor under different copper concentrations through interaction with RNA polymerase(1). CueR forms dimers consisting of three functional domains (a DNA-binding, a dimerisation and a metal-binding domain). The DNA binding domains bind to DNA inverted repeats called CueR boxes with the sequence:
CCTTCCNNNNNNGGAAGG
This box is present at the promoter regions of the copper exporting ATPase CopA, some molybdenum cofactor synthesis genes and the periplasmic copper oxidase protein CueO.(2)
MymT
MymT is a small prokaryotic metallothein discovered in Mycobacterium tuberculosis by Gold et al.(3). It is believed that the protein may help the bacterium survive copper toxicity. MymT is believed to bind up to 7 copper ions but has a preference for 4-6.(3)
Composite
This is how the part is intended to work in vivo:
Copper should induce the expression of the chelator via CueR. The chelator should reduce the free copper concentration therefore reducing it level of expression. This should in theory result in the free copper concentration falling to a low, stable level.
Experience
We cloned pCopA MymT with divergent expressed CueR from a gBlock into the shipping plasmid pSB1C3. E. coli strain MG1655 was transformed using the specific recombinant plasmid and a 5ml culture of a transformed colony was grown overnight.
Absorbance Assay
Separately MymT was cloned from Gblock (BBa_K1980002) into the shipping vector then transferred it into the pBAD, arabinose-inducible commercial expression vector.
We were unable however to detect a drop in solution copper concentration due to copper chelation activity of MymT when expressed from pBAD in MG1655 E. coli strain, using an absorbance assay. Performing a similar procedure with this part also showed no measurable drop in copper concentration.
The assay used was the reagent Bathocuproine disulphonic Acid (BCS). BCS is colourless in the absence of Cu+ but upon exposure to Cu+, BCS forms complexes with Cu+ and absorbs strongly in the 480nm range. In our assays, at concentrations of 50µg/mL, the 480nm absorbance varied linearly with the Cu+ concentration from the detection limit of around 1µM Cu+, to approximately 20µM Cu+. As not only MymT and Csp1 bind copper in the Cu+ form, but the assay also requires singly charged copper, the assays were optimised to include a suitable about of mild reducing agent to ensure reduction of the added CuSO4 (releases Cu2+) to Cu+. After trying L(+)-Ascorbate and DL-Dithiothreitol, and L-Glutathione as candidates, L-Glutathione was selected as it was both mild enough to not damage biological material and efficient at reducing Cu2+. At >2-3 times the concentration of Cu2+ in solution, L-Glutathione had maximum reductive activity against Cu2+.
Modelling by our team suggested that this was because insufficient protein could be expressed to chelate the amount needed to be detectable on the assay (1μM detection limit).
Conclusions
We were unable to demonstrate the ability of this system to buffer free copper concentration we believe that this is because the range of concentrations over which the system operates is below the detection limit of our absorbance assay. Future work should be done to optimise the system so it can operate over a wider range of copper concentrations. This could possibly be done by changing the promoter sequence or the RBS strengths.
References
(1) Danya J. Martell, Chandra P. Joshi, Ahmed Gaballa, Ace George Santiago, Tai-Yen Chen, Won Jung, John D. Helmann, and Peng Chen (2015) “Metalloregulator CueR biases RNA polymerase’s kinetic sampling of dead-end or open complex to repress or activate transcription” Proc Natl Acad Sci U S A. 2015 Nov 3; 112(44): 13467–13472
(2) Yamamoto K, Ishihama A. (2005) “Transcriptional response of Escherichia coli to external copper.” Mol Microbiol. 2005 Apr;56(1):215-27
(3) Ben Gold, Haiteng Deng, Ruslana Bryk, Diana Vargas, David Eliezer, Julia Roberts, Xiuju Jiang, & Carl Nathan (2009) “Identification of a Copper-Binding Metallothionein in Pathogenic Mycobacteria” Nat Chem Biol. 2008 October ; 4(10): 609–616. doi:10.1038/nchembio.109.
Author: Andreas Hadjicharalambous