Difference between revisions of "Part:BBa K1998004"
Tom Collier (Talk | contribs) (→Part Verification and Characterisation) |
Tom Collier (Talk | contribs) (→Part Verification) |
||
| (6 intermediate revisions by the same user not shown) | |||
| Line 22: | Line 22: | ||
===Biology & Literature=== | ===Biology & Literature=== | ||
| − | The first gene in this operon is <i>psbM</i> which acts to stabalise the dimerisation of the PSII complex through it's span over the membrane by way on an α-helix located at the monomer-monomer interface of PSII [1]. It has been shown that it's absence will weaker the dimer interconnection of the core complex and may impair PSII repair, however it is not a necessary factor for PSII biosynthesis [2]. In addition the success of PSII centre assembly decreases when this gene is removed [3]. | + | The first gene in this operon is <i>psbM</i> which acts to stabalise the dimerisation of the PSII complex through it's span over the membrane by way on an α-helix located at the monomer-monomer interface of PSII [1]. It has been shown that it's absence will weaker the dimer interconnection of the core complex and may impair PSII repair, however it is not a necessary factor for PSII biosynthesis [2]. In addition, the success of PSII centre assembly decreases when this gene is removed [3]. |
<br><br> | <br><br> | ||
The <i>psbZ</i> gene is also referred to as <i>ycf9</i> [4]. The gene highly conserved gene amongst photosynthetic species and interacts with the light harvesting antenna in the PSII complex [4], found close to the PSII interface and light harvesting complex II [5]. The two transmembrane helix protein which it encodes [1], has been indicated to result in decreased stability of the both the PSII and Light Harvesting Complex II if deleted [4]. This suggests that it has a role in anchoring these two complexes. | The <i>psbZ</i> gene is also referred to as <i>ycf9</i> [4]. The gene highly conserved gene amongst photosynthetic species and interacts with the light harvesting antenna in the PSII complex [4], found close to the PSII interface and light harvesting complex II [5]. The two transmembrane helix protein which it encodes [1], has been indicated to result in decreased stability of the both the PSII and Light Harvesting Complex II if deleted [4]. This suggests that it has a role in anchoring these two complexes. | ||
| Line 30: | Line 30: | ||
The next subunit protein in the operon is encoded by the <i>psbW</i> gene which is a transmembrane protein, responsible for photo-protection and dimer stabilisation [10]. It has been shown to have a close association with the PSII reaction centre [11]. The absence of <i>psbW</i> affects the formation of the PSII complex [12, 13] and it's absence in transgenic plants have indicated a higher sensitivity to light based stress [14]. | The next subunit protein in the operon is encoded by the <i>psbW</i> gene which is a transmembrane protein, responsible for photo-protection and dimer stabilisation [10]. It has been shown to have a close association with the PSII reaction centre [11]. The absence of <i>psbW</i> affects the formation of the PSII complex [12, 13] and it's absence in transgenic plants have indicated a higher sensitivity to light based stress [14]. | ||
<br><br> | <br><br> | ||
| − | The final gene in this operon encodes <i>psbK</i>, another subunit of PSII involved in maintaining the stability of the complex [15]. It has been shown to be tightly associated with the PSII complex, in particular the CP43 antenna complex [16]. Some destabilisation of the PSII complex can be detected if removed from C. reinhardtii [15] however it is not necessary for the formation of the complex. | + | The final gene in this operon encodes <i>psbK</i>, another subunit of PSII involved in maintaining the stability of the complex [15]. It has been shown to be tightly associated with the PSII complex, in particular the CP43 antenna complex [16]. Some destabilisation of the PSII complex can be detected if removed from <i>C. reinhardtii</i> [15] however it is not necessary for the formation of the complex. |
| − | ===Part Verification and Characterisation=== | + | ===Part Verification=== |
| + | |||
| + | <html><centre><img src=" https://static.igem.org/mediawiki/2016/8/82/T--Macquarie_Australia--PSII_Show_Gel_and_.JPG " height="20%" width="40%"></center></html> | ||
| + | <br><br> | ||
| + | <b>Fig 1.</b> Gel electrophoresis of the Photosystem II parts, here <i>psbMZHWK</i> is shown to be the correct size of 1165bp with a cam backbone of 2,070bp, when compaired to 1kb ladder on the gel. This gel validates all Biobrick parts and backbones to the designed constructs. | ||
| + | |||
| + | ===Part Characterisation=== | ||
<br><br> | <br><br> | ||
<html><centre><img src=" https://static.igem.org/mediawiki/2016/6/69/T--Macquarie_Australia--PSII_MZHWK_Show_Gel_.JPG " height="20%" width="40%"></center></html> | <html><centre><img src=" https://static.igem.org/mediawiki/2016/6/69/T--Macquarie_Australia--PSII_MZHWK_Show_Gel_.JPG " height="20%" width="40%"></center></html> | ||
<br><br> | <br><br> | ||
| − | <b>Fig | + | <b>Fig 2.</b> Protein Gel of fractionated MZHWK expressing <i>E. coli</i>. Lane 2 soluble protein fraction, lane 3 urea washing of pellet and lane 4 insoluble pellet) over expressed. MZHWK is shown here to be the correct molecular size (27kD). Following SDS treatment and boiling this single band remains in high yields suggesting it has an extremely hydrophobic nature. |
<br><br> | <br><br> | ||
| − | This Biobrick construct is a composite part of 5 individual genes. Each gene encodes a protein which range in size from 4 kDa for the smallest to 9 kDa as the largest. When expressed these proteins combined size is 27 kDa, which is exactly the size of the protein produced in E. coli containing this plasmid induced with IPTG as it appeared on our SDS-PAGE protein gel (Fig | + | This Biobrick construct is a composite part of 5 individual genes. Each gene encodes a protein which range in size from 4 kDa for the smallest to 9 kDa as the largest. When expressed these proteins combined size is 27 kDa, which is exactly the size of the protein produced in E. coli containing this plasmid induced with IPTG as it appeared on our SDS-PAGE protein gel (Fig 2). We have a single band, expressing at a very high yield, representing a homogenous complex of the 5 expressed protein. We believe this extremely hydrophobic and stable complex could be membrane associated. |
<br><br> | <br><br> | ||
<html><centre><img src=" https://static.igem.org/mediawiki/2016/2/2c/T--Macquarie_Australia--Green_Test_Tubes_.JPG " height="20%" width="40%"></center></html> | <html><centre><img src=" https://static.igem.org/mediawiki/2016/2/2c/T--Macquarie_Australia--Green_Test_Tubes_.JPG " height="20%" width="40%"></center></html> | ||
<br><br> | <br><br> | ||
| − | <b>Fig | + | <b>Fig 3.</b> Binding of chlorophyll to MZHWK complex as shown by green pellet on left indicating chlorophyll has associated with insoluble pellet fraction (Tube 1, from left). Corresponding soluble fraction is shown in Tube 2. In contrast, chlorophyll does not associate with the Cas9 control cells with the majority of the chlorophyll remaining in the soluble fraction following centrifugation (Tube 4, far right) and is not found associated with insoluble pellet fraction (Tube 3). |
<br><br> | <br><br> | ||
The majority of the complex was found in the insoluble pellet fraction (Fig 1. lane 4). MZHWK was tested by binding chlorophyll via mixing the insoluble membrane pellet fraction with purified chlorophyll, and we observed that the pellet then turns green, while the unrelated inclusion body remains white; the supernatant of the unrelated protein control remains green, while the MZHWK supernatant turns clear. This provides evidence that the MZHWK protein complex binds chlorophyll in vitro. | The majority of the complex was found in the insoluble pellet fraction (Fig 1. lane 4). MZHWK was tested by binding chlorophyll via mixing the insoluble membrane pellet fraction with purified chlorophyll, and we observed that the pellet then turns green, while the unrelated inclusion body remains white; the supernatant of the unrelated protein control remains green, while the MZHWK supernatant turns clear. This provides evidence that the MZHWK protein complex binds chlorophyll in vitro. | ||
| − | |||
| − | |||
| − | |||
| − | |||
===Protein information=== | ===Protein information=== | ||
Latest revision as of 03:40, 21 October 2016
psbMZHWK
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 570
- 1000COMPATIBLE WITH RFC[1000]
Overview
This part is composed of the psbM, psbZ, psbH, psbW and psbK genes. The psbM protein subunit is positioned at the monomer-monomer interface. The psbZ protein controls the interaction of Photosystem II cores with the light-harvesting antenna. The psbH protein is required for stability and assembly of the photosystem II complex. The psbW protein stabilizes dimeric photosytem II. The psbK protein is also required for stability and assembly of Photosystem II.
These parts make up one of the operons in our PSII pathway.
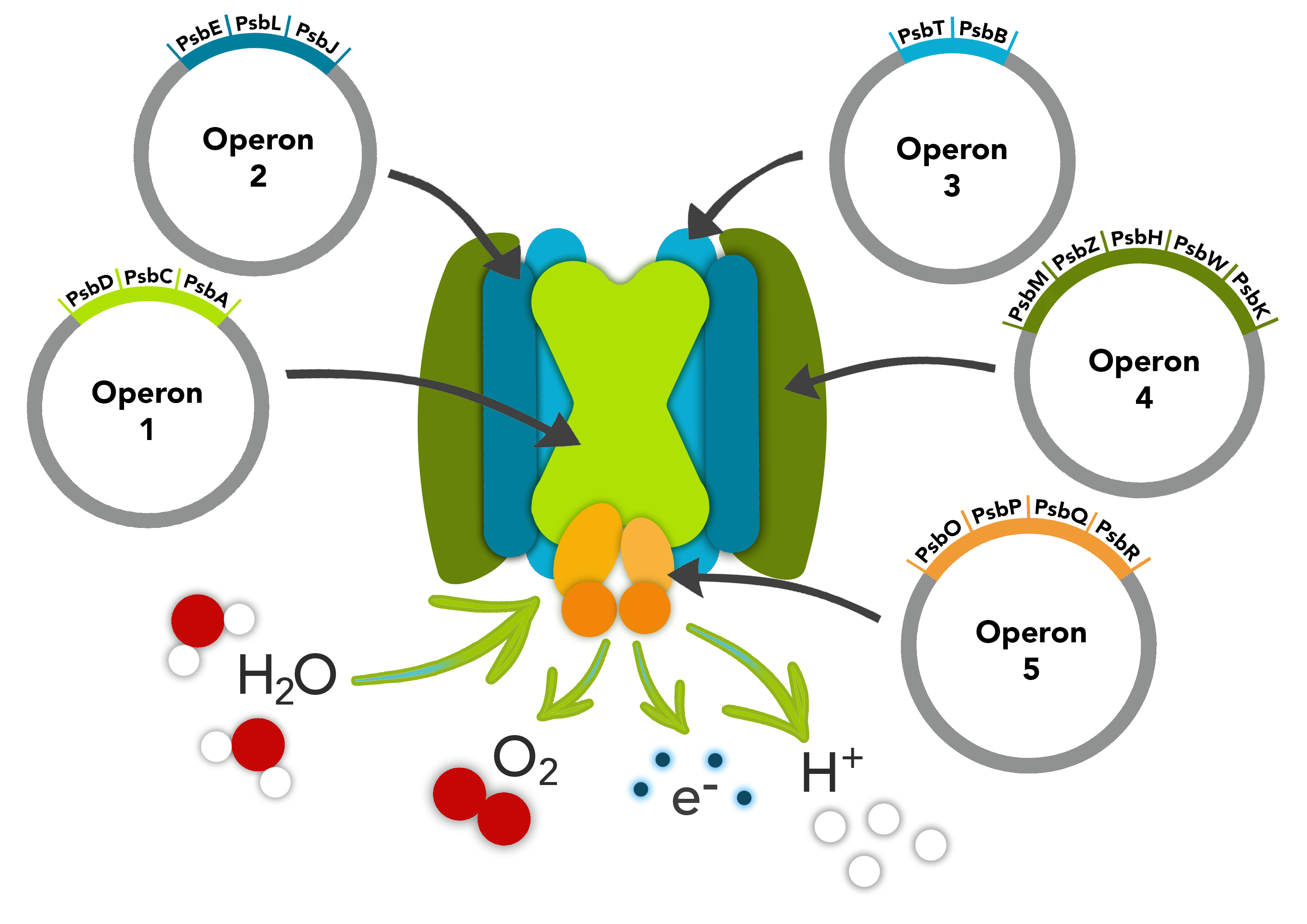
Biology & Literature
The first gene in this operon is psbM which acts to stabalise the dimerisation of the PSII complex through it's span over the membrane by way on an α-helix located at the monomer-monomer interface of PSII [1]. It has been shown that it's absence will weaker the dimer interconnection of the core complex and may impair PSII repair, however it is not a necessary factor for PSII biosynthesis [2]. In addition, the success of PSII centre assembly decreases when this gene is removed [3].
The psbZ gene is also referred to as ycf9 [4]. The gene highly conserved gene amongst photosynthetic species and interacts with the light harvesting antenna in the PSII complex [4], found close to the PSII interface and light harvesting complex II [5]. The two transmembrane helix protein which it encodes [1], has been indicated to result in decreased stability of the both the PSII and Light Harvesting Complex II if deleted [4]. This suggests that it has a role in anchoring these two complexes.
The third gene in this operon, psbH contains a membrane spanning alpha helix [6] and contains multiple phosphorylation sites [7]. Deletion of the psbH complex destabilises the PSII complex and affects the ability of bicarbonate to bind to the complex [8]. In another studying the elimination of the gene in Chlamydomonas reinhardtii, affects the ability of the PSII complex to form, therefore indicating the importance of the psbH gene for the synthesis of Photosystem II [9].
The next subunit protein in the operon is encoded by the psbW gene which is a transmembrane protein, responsible for photo-protection and dimer stabilisation [10]. It has been shown to have a close association with the PSII reaction centre [11]. The absence of psbW affects the formation of the PSII complex [12, 13] and it's absence in transgenic plants have indicated a higher sensitivity to light based stress [14].
The final gene in this operon encodes psbK, another subunit of PSII involved in maintaining the stability of the complex [15]. It has been shown to be tightly associated with the PSII complex, in particular the CP43 antenna complex [16]. Some destabilisation of the PSII complex can be detected if removed from C. reinhardtii [15] however it is not necessary for the formation of the complex.
Part Verification

Fig 1. Gel electrophoresis of the Photosystem II parts, here psbMZHWK is shown to be the correct size of 1165bp with a cam backbone of 2,070bp, when compaired to 1kb ladder on the gel. This gel validates all Biobrick parts and backbones to the designed constructs.
Part Characterisation

Fig 2. Protein Gel of fractionated MZHWK expressing E. coli. Lane 2 soluble protein fraction, lane 3 urea washing of pellet and lane 4 insoluble pellet) over expressed. MZHWK is shown here to be the correct molecular size (27kD). Following SDS treatment and boiling this single band remains in high yields suggesting it has an extremely hydrophobic nature.
This Biobrick construct is a composite part of 5 individual genes. Each gene encodes a protein which range in size from 4 kDa for the smallest to 9 kDa as the largest. When expressed these proteins combined size is 27 kDa, which is exactly the size of the protein produced in E. coli containing this plasmid induced with IPTG as it appeared on our SDS-PAGE protein gel (Fig 2). We have a single band, expressing at a very high yield, representing a homogenous complex of the 5 expressed protein. We believe this extremely hydrophobic and stable complex could be membrane associated.

Fig 3. Binding of chlorophyll to MZHWK complex as shown by green pellet on left indicating chlorophyll has associated with insoluble pellet fraction (Tube 1, from left). Corresponding soluble fraction is shown in Tube 2. In contrast, chlorophyll does not associate with the Cas9 control cells with the majority of the chlorophyll remaining in the soluble fraction following centrifugation (Tube 4, far right) and is not found associated with insoluble pellet fraction (Tube 3).
The majority of the complex was found in the insoluble pellet fraction (Fig 1. lane 4). MZHWK was tested by binding chlorophyll via mixing the insoluble membrane pellet fraction with purified chlorophyll, and we observed that the pellet then turns green, while the unrelated inclusion body remains white; the supernatant of the unrelated protein control remains green, while the MZHWK supernatant turns clear. This provides evidence that the MZHWK protein complex binds chlorophyll in vitro.
Protein information
psbM
mass: 3.76kDa
sequence: MEVNIYGLTATALFIIIPTSFLLILYVKTASTQD
psbZ
mass: 4.56kDa
sequence: MVGVPVVFATPNGWTDNKGAVFSGLSLWLLLVFVVGILNSFVV
psbH
mass: 6.02kDa
sequence: MSEAGKVLPGWGTTVLMAVFILLFAAFLLIILEIYNSSLILDDVSMSWETLAKVS
psbW
mass: 9.2kDa
sequence: MATTVRSEVAKKVAMLSTLPATLAAHPAFALVDERMNGDGTGRPFGVNDPVLGWVLLGVFGTMWAIWFIGQKDLGDFEDADDGLKL
psbK
mass: 5.0kDa
sequence: MTTLALVLAKLPEAYAPFAPIVDVLPVIPVFFILLAFVWQAAVSFR
References
[1] Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S. Architecture of the photosynthetic oxygen-evolving center. Science. 2004 Mar 19;303(5665):1831-8.
[2] Umate P, Schwenkert S, Karbat I, Dal Bosco C, Mlcòchová L, Volz S, Zer H, Herrmann RG, Ohad I, Meurer J. Deletion of PsbM in tobacco alters the QB site properties and the electron flow within photosystem II. Journal of Biological Chemistry. 2007 Mar 30;282(13):9758-67.
[3] Bentley FK, Luo H, Dilbeck P, Burnap RL, Eaton-Rye JJ. Effects of Inactivating psbM and psbT on Photodamage and Assembly of Photosystem II in Synechocystis sp. PCC 6803†. Biochemistry. 2008 Oct 4;47(44):11637-46.
[4] Swiatek M, Kuras R, Sokolenko A, Higgs D, Olive J, Cinque G, Müller B, Eichacker LA, Stern DB, Bassi R, Herrmann RG. The chloroplast gene ycf9 encodes a photosystem II (PSII) core subunit, PsbZ, that participates in PSII supramolecular architecture. The Plant Cell. 2001 Jun 1;13(6):1347-68.
[5] Minagawa J, Takahashi Y. Structure, function and assembly of Photosystem II and its light-harvesting proteins. Photosynthesis research. 2004 Dec 1;82(3):241-63.
[6] Michel HP, Bennett J. Identification of the phosphorylation site of an 8.3 kDa protein from photosystem II of spinach. FEBS letters. 1987 Feb 9;212(1):103-8.
[7] Vener AV, Harms A, Sussman MR, Vierstra RD. Mass spectrometric resolution of reversible protein phosphorylation in photosynthetic membranes ofArabidopsis thaliana. Journal of Biological Chemistry. 2001 Mar 9;276(10):6959-66.
[8] Komenda J, Lupinkova L, Kopecký J. Absence of the psbH gene product destabilizes photosystem II complex and bicarbonate binding on its acceptor side in Synechocystis PCC 6803. European Journal of Biochemistry. 2002 Jan 1;269(2):610-9.
[9] Summer EJ, Schmid VH, Bruns BU, Schmidt GW. Requirement for the H phosphoprotein in photosystem II of Chlamydomonas reinhardtii. Plant physiology. 1997 Apr 1;113(4):1359-68.
[10] Woolhead CA, Mant A, Kim SJ, Robinson C, Rodger A. Conformation of a purified “spontaneously” inserting thylakoid membrane protein precursor in aqueous solvent and detergent micelles. Journal of Biological Chemistry. 2001 May 4;276(18):14607-13.
[11] Shi LX, Schröder WP. Compositional and topological studies of the PsbW protein in spinach thylakoid membrane. Photosynthesis research. 1997 Jul 1;53(1):45-53.
[12] Shi LX, Lorković ZJ, Oelmüller R, Schröder WP. The low molecular mass PsbW protein is involved in the stabilization of the dimeric photosystem II complex in Arabidopsis thaliana. Journal of Biological Chemistry. 2000 Dec 1;275(48):37945-50.
[13] García‐Cerdán JG, Kovács L, Tóth T, Kereïche S, Aseeva E, Boekema EJ, Mamedov F, Funk C, Schröder WP. The PsbW protein stabilizes the supramolecular organization of photosystem II in higher plants. The Plant Journal. 2011 Feb 1;65(3):368-81.
[14] Thidholm E, Shi LX, Schroder W. The PsbW-protien; Its location and involvement in photoinhibition. Science Access. 2001;3(1).
[15] Takahashi Y, Matsumoto H, Goldschmidt-Clermont M, Rochaix JD. Directed disruption of the Chlamydomonas chloroplast psbK gene destabilizes the photosystem II reaction center complex. Plant molecular biology. 1994 Mar 1;24(5):779-88.
[16] Sugimoto I, Takahashi Y. Evidence that the PsbK polypeptide is associated with the photosystem II core antenna complex CP43. Journal of Biological Chemistry. 2003 Nov 7;278(45):45004-10.
